5ETT
 
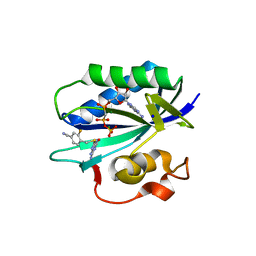 | | S. aureus 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase complexed with AMPCPP and inhibitor at 1.55 angstrom resolution | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 4-[(2-azanyl-6-oxidanylidene-1,9-dihydropurin-8-yl)sulfanylmethyl]-3-fluoranyl-benzenecarbonitrile, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Dennis, M.L, Peat, T.S, Swarbrick, J.D. | | Deposit date: | 2015-11-18 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Basis for the Selective Binding of Inhibitors to 6-Hydroxymethyl-7,8-dihydropterin Pyrophosphokinase from Staphylococcus aureus and Escherichia coli.
J.Med.Chem., 59, 2016
|
|
5ETQ
 
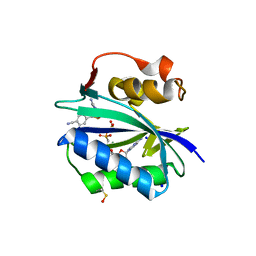 | | S. aureus 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase complexed with AMPCPP and inhibitor at 1.96 angstrom resolution | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 4-{[(2-amino-6-oxo-6,9-dihydro-1H-purin-8-yl)sulfanyl]methyl}benzonitrile, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Dennis, M.L, Peat, T.S, Swarbrick, J.D. | | Deposit date: | 2015-11-18 | | Release date: | 2016-05-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural Basis for the Selective Binding of Inhibitors to 6-Hydroxymethyl-7,8-dihydropterin Pyrophosphokinase from Staphylococcus aureus and Escherichia coli.
J.Med.Chem., 59, 2016
|
|
5ETP
 
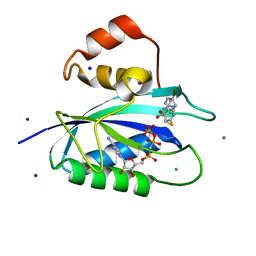 | | E. coli 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase complexed with AMPCPP and inhibitor at 1.05 angstrom resolution | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 2-azanyl-8-[2-(4-bromophenyl)-2-oxidanylidene-ethyl]sulfanyl-1,9-dihydropurin-6-one, CALCIUM ION, ... | | Authors: | Dennis, M.L, Peat, T.S, Swarbrick, J.D. | | Deposit date: | 2015-11-17 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural Basis for the Selective Binding of Inhibitors to 6-Hydroxymethyl-7,8-dihydropterin Pyrophosphokinase from Staphylococcus aureus and Escherichia coli.
J.Med.Chem., 59, 2016
|
|
5ETR
 
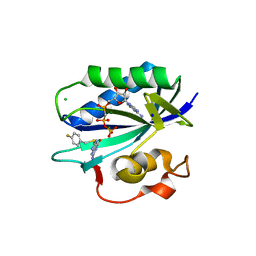 | | S. aureus 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase complexed with AMPCPP and inhibitor at 1.32 angstrom resolution | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 2-azanyl-8-[(4-fluorophenyl)methylsulfanyl]-1,7-dihydropurin-6-one, CHLORIDE ION, ... | | Authors: | Dennis, M.L, Peat, T.S, Swarbrick, J.D. | | Deposit date: | 2015-11-18 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural Basis for the Selective Binding of Inhibitors to 6-Hydroxymethyl-7,8-dihydropterin Pyrophosphokinase from Staphylococcus aureus and Escherichia coli.
J.Med.Chem., 59, 2016
|
|
5F3H
 
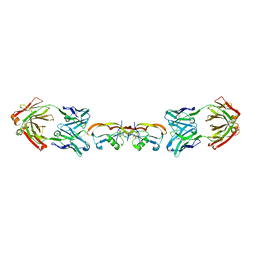 | | Structure of myostatin in complex with humanized RK35 antibody | | Descriptor: | Growth/differentiation factor 8, humanized RK35 antibody heavy chain, humanized RK35 antibody light chain | | Authors: | Parris, K.D, Mosyak, L. | | Deposit date: | 2015-12-02 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Beyond CDR-grafting: Structure-guided humanization of framework and CDR regions of an anti-myostatin antibody.
Mabs, 8, 2016
|
|
5F4R
 
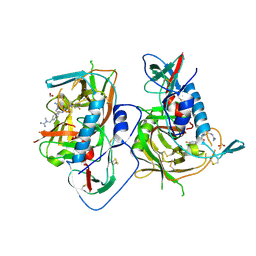 | | HIV-1 gp120 complex with BNW-IV-147 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENVELOPE GLYCOPROTEIN GP120 of HIV-1 clade C, FORMIC ACID, ... | | Authors: | Liang, S, Hendrickson, W.A. | | Deposit date: | 2015-12-03 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Small-Molecule CD4-Mimics: Structure-Based Optimization of HIV-1 Entry Inhibition.
ACS Med Chem Lett, 7, 2016
|
|
5G5J
 
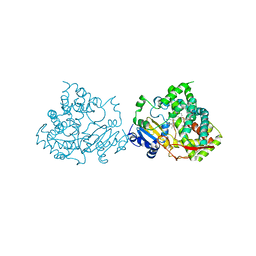 | | Crystal structure of human CYP3A4 bound to metformin | | Descriptor: | CYTOCHROME P450 3A4, Metformin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioukova, I. | | Deposit date: | 2016-05-25 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Heme Binding Biguanides Target Cytochrome P450-Dependent Cancer Cell Mitochondria.
Cell Chem Biol, 24, 2017
|
|
8QZN
 
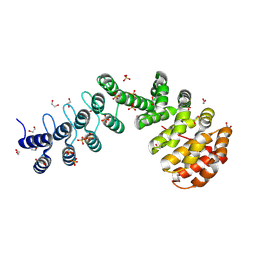 | | DARPin-Armadillo fusion protein with a selected Tyrosin pocket binding to the fused target peptide | | Descriptor: | 1,2-ETHANEDIOL, DARPin-Armadillo fusion protein with Tyrosin pocket and linked target peptide, SULFATE ION | | Authors: | Stark, Y, Menard, F, Jeliazkov, J, Ernst, P, Chembath, A, Ashraf, M, Hine, A, Plueckthun, A. | | Deposit date: | 2023-10-27 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Modular binder technology by NGS-aided, high-resolution selection in yeast of designed armadillo modules.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
3PMH
 
 | |
3PIQ
 
 | | Crystal structure of human 2909 Fab, a quaternary structure-specific antibody against HIV-1 | | Descriptor: | Human monoclonal antibody 2909 Fab heavy chain, Human monoclonal antibody 2909 Fab light chain | | Authors: | Changela, A, Gorny, M.K, Zolla-Pazner, S, Kwong, P.D. | | Deposit date: | 2010-11-07 | | Release date: | 2011-01-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.325 Å) | | Cite: | Crystal Structure of Human Antibody 2909 Reveals Conserved Features of Quaternary Structure-Specific Antibodies That Potently Neutralize HIV-1.
J.Virol., 85, 2011
|
|
3Q6W
 
 | | Structure of dually-phosphorylated MET receptor kinase in complex with an MK-2461 analog with specificity for the activated receptor | | Descriptor: | 3-{5-oxo-3-[1-(piperidin-4-yl)-1H-pyrazol-4-yl]-5H-benzo[4,5]cyclohepta[1,2-b]pyridin-7-yl}-N-(pyridin-2-ylmethyl)propanamide, Hepatocyte growth factor receptor | | Authors: | Soisson, S.M, Rickert, K.W, Patel, S.B, Lumb, K.J. | | Deposit date: | 2011-01-03 | | Release date: | 2011-01-19 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for selective small molecule kinase inhibition of activated c-Met.
J.Biol.Chem., 286, 2011
|
|
8SMT
 
 | | Crystal structure of antibody WRAIR-2134 in complex with SARS-CoV-2 receptor binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, WRAIR-2134 Fab heavy chain, ... | | Authors: | Sankhala, R.S, Jensen, J.L, Joyce, M.G. | | Deposit date: | 2023-04-26 | | Release date: | 2023-06-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Antibody targeting of conserved sites of vulnerability on the SARS-CoV-2 spike receptor-binding domain.
Structure, 32, 2024
|
|
3S6W
 
 | | Crystal structure of Tudor domain of human TDRD3 | | Descriptor: | ISOPROPYL ALCOHOL, Tudor domain-containing protein 3 | | Authors: | Liu, H.P, Xu, R.M. | | Deposit date: | 2011-05-26 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of TDRD3 and methyl-arginine binding characterization of TDRD3, SMN and SPF30
Plos One, 7, 2012
|
|
1R1C
 
 | | PSEUDOMONAS AERUGINOSA W48F/Y72F/H83Q/Y108W-AZURIN RE(PHEN)(CO)3(HIS107) | | Descriptor: | (1,10 PHENANTHROLINE)-(TRI-CARBON MONOXIDE) RHENIUM (I), Azurin, COPPER (I) ION | | Authors: | Miller, J.E, Gradinaru, C, Crane, B.R, Di Bilio, A.J. | | Deposit date: | 2003-09-23 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Spectroscopy and reactivity of a photogenerated tryptophan radical in a structurally defined protein environment
J.Am.Chem.Soc., 125, 2003
|
|
8S9D
 
 | |
8S97
 
 | |
8SMQ
 
 | | Crystal Structure of the N-terminal Domain of the Cryptic Surface Protein (CD630_25440) from Clostridium difficile. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Kiryukhina, O, Wawrzak, Z, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2023-04-26 | | Release date: | 2023-05-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein target highlights in CASP15: Analysis of models by structure providers.
Proteins, 91, 2023
|
|
3SE9
 
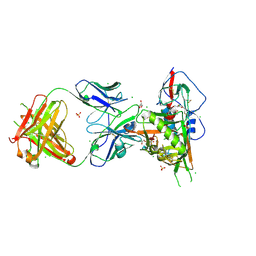 | | Crystal structure of broadly and potently neutralizing antibody VRC-PG04 in complex with HIV-1 gp120 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kwong, P.D, Zhou, T. | | Deposit date: | 2011-06-10 | | Release date: | 2011-08-10 | | Last modified: | 2021-04-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing.
Science, 333, 2011
|
|
8S9F
 
 | |
8S93
 
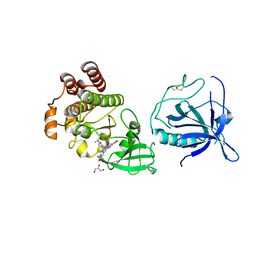 | | Crystal structure of the PH-TH/kinase complex of Bruton's tyrosine kinase | | Descriptor: | 2-[3'-(hydroxymethyl)-1-methyl-5-({5-[(2S)-2-methyl-4-(oxetan-3-yl)piperazin-1-yl]pyridin-2-yl}amino)-6-oxo[1,6-dihydro[3,4'-bipyridine]]-2'-yl]-7,7-dimethyl-3,4,7,8-tetrahydro-2H-cyclopenta[4,5]pyrrolo[1,2-a]pyrazin-1(6H)-one, GLYCEROL, Tyrosine-protein kinase BTK, ... | | Authors: | Lin, D.Y, Andreotti, A.H. | | Deposit date: | 2023-03-27 | | Release date: | 2023-08-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational heterogeneity of the BTK PHTH domain drives multiple regulatory states.
Elife, 12, 2024
|
|
8SXR
 
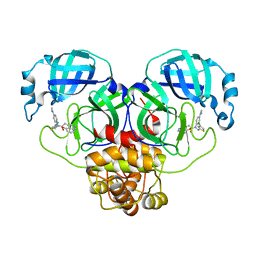 | | Crystal structure of SARS-CoV-2 Mpro with C5a | | Descriptor: | 3C-like proteinase nsp5, N-[(4-chlorothiophen-2-yl)methyl]-N-[4-(dimethylamino)phenyl]-2-(5-hydroxyisoquinolin-4-yl)acetamide | | Authors: | Worrall, L.J, Kenward, C, Lee, J, Strynadka, N.C.J. | | Deposit date: | 2023-05-23 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.114 Å) | | Cite: | A novel class of broad-spectrum active-site-directed 3C-like protease inhibitors with nanomolar antiviral activity against highly immune-evasive SARS-CoV-2 Omicron subvariants.
Emerg Microbes Infect, 12, 2023
|
|
3QYB
 
 | |
8SMI
 
 | | Crystal structure of antibody WRAIR-2123 in complex with SARS-CoV-2 receptor binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, WRAIR-2123 Fab heavy chain, ... | | Authors: | Sankhala, R.S, Jensen, J.L, Joyce, M.G. | | Deposit date: | 2023-04-26 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Antibody targeting of conserved sites of vulnerability on the SARS-CoV-2 spike receptor-binding domain.
Structure, 32, 2024
|
|
8SGU
 
 | | Crystal structure of the SARS-CoV-2 receptor binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Sankhala, R.S, Jensen, J.L, Joyce, M.G. | | Deposit date: | 2023-04-13 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Antibody targeting of conserved sites of vulnerability on the SARS-CoV-2 spike receptor-binding domain.
Structure, 32, 2024
|
|
8SMX
 
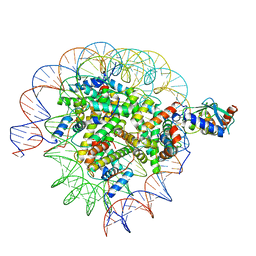 | | Cryo-EM structure of the human nucleosome core particle in complex with RNF168 and UbcH5c~Ub (UbcH5c chemically conjugated to histone H2A. No density for Ub.) (class 2) | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-04-26 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
