1MBE
 
 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 1 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-07-31 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
1MBG
 
 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 2 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-07-31 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
1MBJ
 
 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 3 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-07-31 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
1MBK
 
 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 3 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-07-31 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
4GHQ
 
 | | Crystal structure of EV71 3C proteinase | | Descriptor: | 3C proteinase | | Authors: | Chen, C, Wu, C, Cai, Q, Li, N, Peng, X, Cai, Y, Yin, K, Chen, X, Wang, X, Zhang, R, Liu, L, Chen, S, Li, J, Lin, T. | | Deposit date: | 2012-08-08 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of Enterovirus 71 3C proteinase (strain E2004104-TW-CDC) and its complex with rupintrivir
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1MBF
 
 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 1 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-07-31 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
4H5Y
 
 | | High-resolution crystal structure of Legionella pneumophila LidA (60-594) | | Descriptor: | LidA protein, substrate of the Dot/Icm system | | Authors: | An, X, Ye, S, Liu, Y, Zheng, X, Zhang, R. | | Deposit date: | 2012-09-19 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of LidA, a translocated substrate of the Legionella pneumophila type IV secretion system.
Protein Cell, 4, 2013
|
|
1P53
 
 | | The Crystal Structure of ICAM-1 D3-D5 fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Intercellular adhesion molecule-1 | | Authors: | Yang, Y, Jun, C.D, Liu, J.H, Zhang, R, Jochimiak, A, Springer, T.A, Wang, J.H. | | Deposit date: | 2003-04-24 | | Release date: | 2004-05-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structural basis for dimerization of ICAM-1 on the cell surface.
Mol.Cell, 14, 2004
|
|
3CQV
 
 | | Crystal structure of Reverb beta in complex with heme | | Descriptor: | Nuclear receptor subfamily 1 group D member 2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Xu, X, Dong, A, Pardee, K.I, Reinking, J, Krause, H, Schuetz, A, Zhang, R, Cui, H, Edwards, A, Arrowsmith, C.H, Weigelt, J, Bountra, C, Savchenko, A, Botchkarev, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-04-03 | | Release date: | 2008-08-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis of gas-responsive transcription by the human nuclear hormone receptor REV-ERBbeta.
Plos Biol., 7, 2009
|
|
3D6K
 
 | | The crystal structure of a putative aminotransferase from Corynebacterium diphtheriae | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Putative aminotransferase, ... | | Authors: | Tan, K, Zhang, R, Duggan, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-19 | | Release date: | 2008-07-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of a putative aminotransferase from Corynebacterium diphtheriae
To be Published
|
|
1V4A
 
 | | Structure of the N-terminal Domain of Escherichia coli Glutamine Synthetase adenylyltransferase | | Descriptor: | Glutamate-ammonia-ligase adenylyltransferase | | Authors: | Xu, Y, Zhang, R, Joachimiak, A, Carr, P.D, Ollis, D.L, Vasudevan, S.G. | | Deposit date: | 2003-11-12 | | Release date: | 2004-07-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the n-terminal domain of Escherichia coli glutamine synthetase adenylyltransferase
Structure, 12, 2004
|
|
3KSN
 
 | | Crystal structure of the lipoprotein localization factor, LolA | | Descriptor: | Outer-membrane lipoprotein carrier protein | | Authors: | Ahmadpour, F, Gloyd, M, Guarne, A, Stewart, G, Pathania, R, Brown, E.D. | | Deposit date: | 2009-11-23 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the lipoprotein localization factor, LolA
To be Published
|
|
1EXZ
 
 | | STRUCTURE OF STEM CELL FACTOR | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, SAMARIUM (III) ION, ... | | Authors: | Zhang, Z, Zhang, R, Joachimiak, A, Schlessinger, J, Kong, X. | | Deposit date: | 2000-05-05 | | Release date: | 2000-07-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human stem cell factor: implication for stem cell factor receptor dimerization and activation.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
2QMW
 
 | | The crystal structure of the prephenate dehydratase (PDT) from Staphylococcus aureus subsp. aureus Mu50 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tan, K, Zhang, R, Li, H, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-17 | | Release date: | 2007-08-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of open (R) and close (T) states of prephenate dehydratase (PDT) - implication of allosteric regulation by L-phenylalanine.
J.Struct.Biol., 162, 2008
|
|
5HFM
 
 | | Gp41-targeting HIV-1 fusion inhibitors with hook-like Ile-Asp-Leu tail | | Descriptor: | Envelope glycoprotein gp160,gp41 CHR region, TRIS(HYDROXYETHYL)AMINOMETHANE | | Authors: | Zhu, Y, Ye, S, Zhang, R. | | Deposit date: | 2016-01-07 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Rational improvement of gp41-targeting HIV-1 fusion inhibitors: an innovatively designed Ile-Asp-Leu tail with alternative conformations
Sci Rep, 6, 2016
|
|
5HFL
 
 | |
3HXT
 
 | | Structure of human MTHFS | | Descriptor: | 5-formyltetrahydrofolate cyclo-ligase, MAGNESIUM ION, NICKEL (II) ION | | Authors: | Wu, D, Li, Y, Song, G, Cheng, C, Zhang, R, Joachimiak, A, Shaw, N, Liu, Z.-J. | | Deposit date: | 2009-06-22 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the inhibition of human 5,10-methenyltetrahydrofolate synthetase by N10-substituted folate analogues
Cancer Res., 69, 2009
|
|
1YOZ
 
 | | Predicted coding region AF0941 from Archaeoglobus fulgidus | | Descriptor: | Hypothetical protein AF0941 | | Authors: | Lunin, V.V, Zhang, R, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-01-28 | | Release date: | 2005-02-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of predicted coding region AF0941 from Archaeoglobus fulgidus
To be Published
|
|
4RF9
 
 | | Crystal structure of double-domain arginine kinase from Anthopleura japonicas in complex with L-arginine and ATPgS | | Descriptor: | ACETATE ION, ARGININE, Arginine kinase, ... | | Authors: | Wang, Z, Qiao, Z, Ye, S, Zhang, R. | | Deposit date: | 2014-09-25 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of a double-domain phosphagen kinase reveals an asymmetric arrangement of the tandem domains.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2BBJ
 
 | | Crystal structure of the CorA Mg2+ transporter | | Descriptor: | divalent cation transport-related protein | | Authors: | Lunin, V.V, Dobrovetsky, E, Khutoreskaya, G, Zhang, R, Joachimiak, A, Bochkarev, A, Maguire, M.E, Edwards, A.M, Koth, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-10-17 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Crystal structure of the CorA Mg2+ transporter
Nature, 440, 2006
|
|
3ZO9
 
 | | The structure of Trehalose Synthase (TreS) of Mycobacterium smegmatis | | Descriptor: | CALCIUM ION, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Caner, S, Nguyen, N, Aguda, A, Zhang, R, Pan, Y.T, Withers, S.G, Brayer, G.D. | | Deposit date: | 2013-02-21 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The Structure of the Mycobacterium Smegmatis Trehalose Synthase Reveals an Unusual Active Site Configuration and Acarbose-Binding Mode.
Glycobiology, 23, 2013
|
|
4RF8
 
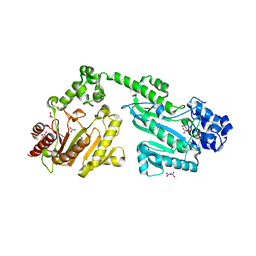 | | Crystal structure of double-domain arginine kinase from Anthopleura japonicas in complex with ADP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, Arginine kinase, ... | | Authors: | Wang, Z, Qiao, Z, Ye, S, Zhang, R. | | Deposit date: | 2014-09-25 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structure of a double-domain phosphagen kinase reveals an asymmetric arrangement of the tandem domains.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3GE2
 
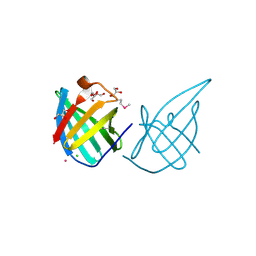 | | Crystal structure of putative lipoprotein SP_0198 from Streptococcus pneumoniae | | Descriptor: | CHLORIDE ION, GLYCEROL, Lipoprotein, ... | | Authors: | Kim, Y, Zhang, R, Joachimiak, G, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-02-24 | | Release date: | 2009-03-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Crystal Structure of Putative Lipoprotein SP_0198 from Streptococcus pneumoniae
To be Published
|
|
1W8I
 
 | | The Structure of gene product af1683 from Archaeoglobus fulgidus. | | Descriptor: | PUTATIVE VAPC RIBONUCLEASE AF_1683 | | Authors: | Midwest Center for Structural Genomics (MCSG), Cuff, M.E, Zhang, R, Ginell, S.L, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A. | | Deposit date: | 2004-09-22 | | Release date: | 2004-11-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of Gene Product Af1683 from Archaeoglobus Fulgidus
To be Published
|
|
8WB1
 
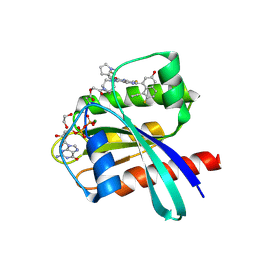 | | Crystal Structure of small molecule KN2H covalently bound to K-Ras(G12D) | | Descriptor: | 1,2-ETHANEDIOL, 1-[(1~{S},5~{R})-3-[7-(8-ethynyl-7-fluoranyl-3-oxidanyl-naphthalen-1-yl)-8-fluoranyl-2-[[(2~{R},8~{S})-2-fluoranyl-1,2,3,5,6,7-hexahydropyrrolizin-8-yl]methoxy]pyrido[4,3-d]pyrimidin-4-yl]-3,8-diazabicyclo[3.2.1]octan-8-yl]ethanone, GTPase KRas, ... | | Authors: | Zhou, L, Zhang, R, Shuai, T.B. | | Deposit date: | 2023-09-08 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal Structure of small molecule KN2H covalently bound to K-Ras(G12D)
To Be Published
|
|
