3QKR
 
 | | Mre11 Rad50 binding domain bound to Rad50 | | Descriptor: | DNA double-strand break repair protein mre11, DNA double-strand break repair rad50 ATPase, PHOSPHATE ION | | Authors: | Williams, G.J, Williams, R.S, Arvai, A, Moncalian, G, Tainer, J.A. | | Deposit date: | 2011-02-01 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | ABC ATPase signature helices in Rad50 link nucleotide state to Mre11 interface for DNA repair.
Nat.Struct.Mol.Biol., 18, 2011
|
|
4NCH
 
 | | Crystal Structure of Pyrococcus furiosis Rad50 L802W mutation | | Descriptor: | DNA double-strand break repair Rad50 ATPase, SULFATE ION | | Authors: | Classen, S, Williams, G.J, Arvai, A.S, Williams, R.S. | | Deposit date: | 2013-10-24 | | Release date: | 2014-03-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | ATP-driven Rad50 conformations regulate DNA tethering, end resection, and ATM checkpoint signaling.
Embo J., 33, 2014
|
|
4NCK
 
 | | Crystal Structure of Pyrococcus furiosis Rad50 R797G mutation | | Descriptor: | CHLORIDE ION, DNA double-strand break repair Rad50 ATPase, MAGNESIUM ION, ... | | Authors: | Classen, S, Williams, G.J, Arvai, A.S, Williams, R.S. | | Deposit date: | 2013-10-24 | | Release date: | 2014-03-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | ATP-driven Rad50 conformations regulate DNA tethering, end resection, and ATM checkpoint signaling.
Embo J., 33, 2014
|
|
4NCI
 
 | | Crystal Structure of Pyrococcus furiosis Rad50 R805E mutation | | Descriptor: | DNA double-strand break repair Rad50 ATPase | | Authors: | Classen, S, Williams, G.J, Arvai, A.S, Williams, R.S. | | Deposit date: | 2013-10-24 | | Release date: | 2014-03-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | ATP-driven Rad50 conformations regulate DNA tethering, end resection, and ATM checkpoint signaling.
Embo J., 33, 2014
|
|
4NCJ
 
 | | Crystal Structure of Pyrococcus furiosis Rad50 R805E mutation with ADP Beryllium Flouride | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA double-strand break repair Rad50 ATPase, ... | | Authors: | Classen, S, Williams, G.J, Arvai, A.S, Williams, R.S. | | Deposit date: | 2013-10-24 | | Release date: | 2014-03-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ATP-driven Rad50 conformations regulate DNA tethering, end resection, and ATM checkpoint signaling.
Embo J., 33, 2014
|
|
1YJ5
 
 | | Molecular architecture of mammalian polynucleotide kinase, a DNA repair enzyme | | Descriptor: | 5' polynucleotide kinase-3' phosphatase FHA domain, 5' polynucleotide kinase-3' phosphatase catalytic domain, SULFATE ION | | Authors: | Bernstein, N.K, Williams, R.S, Rakovszky, M.L, Cui, D, Green, R, Karimi-Busheri, F, Mani, R.S, Galicia, S, Koch, C.A, Cass, C.E, Durocher, D, Weinfeld, M, Glover, J.N.M. | | Deposit date: | 2005-01-13 | | Release date: | 2005-03-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The molecular architecture of the mammalian DNA repair enzyme, polynucleotide kinase.
Mol.Cell, 17, 2005
|
|
1YJM
 
 | | Crystal structure of the FHA domain of mouse polynucleotide kinase in complex with an XRCC4-derived phosphopeptide. | | Descriptor: | 12-mer peptide from DNA-repair protein XRCC4, Polynucleotide 5'-hydroxyl-kinase | | Authors: | Bernstein, N.K, Williams, R.S, Rakovszky, M.L, Cui, D, Green, R, Karimi-Busheri, F, Mani, R.S, Galicia, S, Koch, C.A, Cass, C.E, Durocher, D, Weinfeld, M, Glover, J.N.M. | | Deposit date: | 2005-01-14 | | Release date: | 2005-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The molecular architecture of the mammalian DNA repair enzyme, polynucleotide kinase.
Mol.Cell, 17, 2005
|
|
4J5S
 
 | | TARG1 (C6orf130), Terminal ADP-ribose Glycohydrolase 1 ADP-ribose complex | | Descriptor: | 1,2-ETHANEDIOL, 1,3,2-DIOXABOROLAN-2-OL, BORATE ION, ... | | Authors: | Schellenberg, M.J, Appel, C.D, Krahn, J, Williams, R.S. | | Deposit date: | 2013-02-09 | | Release date: | 2013-03-27 | | Last modified: | 2013-07-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease.
Embo J., 32, 2013
|
|
4J5Q
 
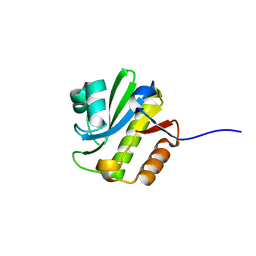 | | TARG1 (C6orf130), Terminal ADP-ribose Glycohydrolase 1, apo structure | | Descriptor: | O-acetyl-ADP-ribose deacetylase 1 | | Authors: | Schellenberg, M.J, Appel, C.D, Krahn, J, Williams, R.S. | | Deposit date: | 2013-02-09 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease.
Embo J., 32, 2013
|
|
4J5R
 
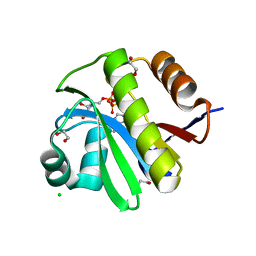 | | TARG1 (C6orf130), Terminal ADP-ribose Glycohydrolase 1 bound to ADP-HPD | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-[(S)-{[(S)-{[(2R,3R,4S)-3,4-DIHYDROXYPYRROLIDIN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]ADENOSINE, CHLORIDE ION, ... | | Authors: | Schellenberg, M.J, Appel, C.D, Krahn, J, Williams, R.S. | | Deposit date: | 2013-02-09 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease.
Embo J., 32, 2013
|
|
4NDF
 
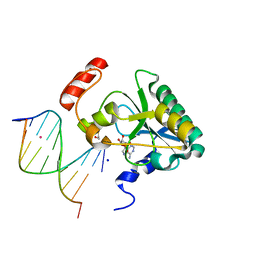 | | Human Aprataxin (Aptx) bound to RNA-DNA, AMP, and Zn - product complex | | Descriptor: | 5'-D(*GP*AP*AP*TP*CP*AP*TP*AP*AP*C)-3', 5'-R(P*G)-D(P*TP*TP*AP*TP*GP*AP*TP*TP*C)-3', ADENOSINE MONOPHOSPHATE, ... | | Authors: | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | Deposit date: | 2013-10-26 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.944 Å) | | Cite: | Aprataxin resolves adenylated RNA-DNA junctions to maintain genome integrity.
Nature, 506, 2013
|
|
4NDI
 
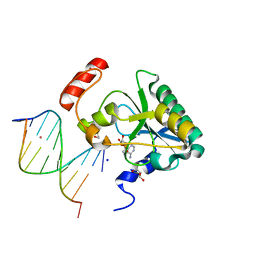 | | Human Aprataxin (Aptx) AOA1 variant K197Q bound to RNA-DNA, AMP, and Zn - product complex | | Descriptor: | 5'-D(*GP*AP*AP*TP*CP*AP*TP*AP*AP*C)-3', 5'-R(P*G)-D(P*TP*TP*AP*TP*GP*AP*TP*TP*C)-3', ADENOSINE MONOPHOSPHATE, ... | | Authors: | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | Deposit date: | 2013-10-26 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Aprataxin resolves adenylated RNA-DNA junctions to maintain genome integrity.
Nature, 506, 2013
|
|
4NDH
 
 | | Human Aprataxin (Aptx) bound to DNA, AMP, and Zn - product complex | | Descriptor: | 5'-D(P*GP*TP*TP*CP*TP*AP*GP*AP*AP*C)-3', ADENOSINE MONOPHOSPHATE, Aprataxin, ... | | Authors: | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | Deposit date: | 2013-10-26 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Aprataxin resolves adenylated RNA-DNA junctions to maintain genome integrity.
Nature, 506, 2013
|
|
4NDG
 
 | | Human Aprataxin (Aptx) bound to RNA-DNA and Zn - adenosine vanadate transition state mimic complex | | Descriptor: | 5'-D(*GP*AP*AP*TP*CP*AP*TP*AP*AP*C)-3', 5'-R(P*G)-D(P*TP*TP*AP*TP*GP*AP*TP*TP*C)-3', Aprataxin, ... | | Authors: | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | Deposit date: | 2013-10-26 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.541 Å) | | Cite: | Aprataxin resolves adenylated RNA-DNA junctions to maintain genome integrity.
Nature, 506, 2013
|
|
6Q01
 
 | | TDP2 UBA Domain Bound to Ubiquitin at 0.85 Angstroms Resolution, Crystal Form 2 | | Descriptor: | 1,2-ETHANEDIOL, BENZOIC ACID, MAGNESIUM ION, ... | | Authors: | Schellenberg, M.J, Krahn, J.M, Williams, R.S. | | Deposit date: | 2019-08-01 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (0.851 Å) | | Cite: | Ubiquitin stimulated reversal of topoisomerase 2 DNA-protein crosslinks by TDP2.
Nucleic Acids Res., 48, 2020
|
|
6Q1V
 
 | | Human DNA Ligase 1 (E592R) Bound to an Adenylated, hydroxyl terminated DNA nick | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Schellenberg, M.J, Williams, R.S, Tumbale, P.S, Riccio, A.A. | | Deposit date: | 2019-08-06 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Two-tiered enforcement of high-fidelity DNA ligation.
Nat Commun, 10, 2019
|
|
7N3Z
 
 | | Crystal Structure of Saccharomyces cerevisiae Apn2 Catalytic Domain | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA-(apurinic or apyrimidinic site) endonuclease 2, ... | | Authors: | Wojtaszek, J.L, Wallace, B.D, Williams, R.S. | | Deposit date: | 2021-06-02 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Molecular basis for processing of topoisomerase 1-triggered DNA damage by Apn2/APE2.
Cell Rep, 41, 2022
|
|
7N3Y
 
 | | Crystal Structure of Saccharomyces cerevisiae Apn2 Catalytic Domain E59Q/D222N Mutant in Complex with DNA | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CITRIC ACID, ... | | Authors: | Wojtaszek, J.L, Krahn, J, Wallace, B.D, Williams, R.S. | | Deposit date: | 2021-06-02 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Molecular basis for processing of topoisomerase 1-triggered DNA damage by Apn2/APE2.
Cell Rep, 41, 2022
|
|
6WCD
 
 | | Crystal Structure of Xenopus laevis APE2 Catalytic Domain | | Descriptor: | 1,2-ETHANEDIOL, 1,4-BUTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Wojtaszek, J.L, Wallace, B.D, Williams, R.S. | | Deposit date: | 2020-03-30 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Endogenous DNA 3' Blocks Are Vulnerabilities for BRCA1 and BRCA2 Deficiency and Are Reversed by the APE2 Nuclease.
Mol.Cell, 78, 2020
|
|
2R1Z
 
 | |
4PUQ
 
 | | Mus Musculus Tdp2 reaction product complex with 5'-phosphorylated RNA/DNA, glycerol, and Mg2+ | | Descriptor: | ACETATE ION, DNA/RNA hybrid, GLYCEROL, ... | | Authors: | Schellenberg, M.J, Williams, R.S. | | Deposit date: | 2014-03-13 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Proteolytic Degradation of Topoisomerase II (Top2) Enables the Processing of Top2DNA and Top2RNA Covalent Complexes by Tyrosyl-DNA-Phosphodiesterase 2 (TDP2).
J.Biol.Chem., 289, 2014
|
|
4GZ0
 
 | |
4GZ1
 
 | |
4GYZ
 
 | | Mus Musculus Tdp2 Bound to dAMP and Mg2+ | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Schellenberg, M.J, Williams, R.S. | | Deposit date: | 2012-09-05 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.556 Å) | | Cite: | Mechanism of repair of 5'-topoisomerase II-DNA adducts by mammalian tyrosyl-DNA phosphodiesterase 2.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4GZ2
 
 | | Mus Musculus Tdp2 excluded ssDNA complex | | Descriptor: | DNA (5'-D(*CP*AP*TP*CP*CP*GP*AP*AP*TP*TP*CP*G)-3'), FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Schellenberg, M.J, Williams, R.S. | | Deposit date: | 2012-09-05 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism of repair of 5'-topoisomerase II-DNA adducts by mammalian tyrosyl-DNA phosphodiesterase 2.
Nat.Struct.Mol.Biol., 19, 2012
|
|
