6A5Y
 
 | | Crystal structure of human FXR/RXR-LBD heterodimer bound to HNC143 and 9cRA and SRC1 | | Descriptor: | (9cis)-retinoic acid, 2-[2-[[3-[2,6-bis(chloranyl)phenyl]-5-cyclopropyl-1,2-oxazol-4-yl]methoxy]-6-azaspiro[3.4]octan-6-yl]-1,3-benzothiazole-6-carboxylic acid, Bile acid receptor, ... | | Authors: | Wang, N, Liu, J. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ligand binding and heterodimerization with retinoid X receptor alpha (RXR alpha ) induce farnesoid X receptor (FXR) conformational changes affecting coactivator binding
J. Biol. Chem., 293, 2018
|
|
6A60
 
 | | Crystal structure of human FXR/RXR-LBD heterodimer bound to GW4064 and 9cRA and SRC1 | | Descriptor: | (9cis)-retinoic acid, 3-[(E)-2-(2-chloro-4-{[3-(2,6-dichlorophenyl)-5-(1-methylethyl)isoxazol-4-yl]methoxy}phenyl)ethenyl]benzoic acid, Bile acid receptor, ... | | Authors: | Wang, N, Liu, J. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Ligand binding and heterodimerization with retinoid X receptor alpha (RXR alpha ) induce farnesoid X receptor (FXR) conformational changes affecting coactivator binding
J. Biol. Chem., 293, 2018
|
|
6A5X
 
 | | FXR-LBD with HNC180 and SRC1 | | Descriptor: | 2-[(1R,5S)-9-[[3-[2,6-bis(chloranyl)phenyl]-5-cyclopropyl-1,2-oxazol-4-yl]methoxy]-3-azabicyclo[3.3.1]nonan-3-yl]-1,3-benzothiazole-6-carboxylic acid, Bile acid receptor, Nuclear receptor coactivator 1, ... | | Authors: | Wang, N, Liu, J. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Ligand binding and heterodimerization with retinoid X receptor alpha (RXR alpha ) induce farnesoid X receptor (FXR) conformational changes affecting coactivator binding
J. Biol. Chem., 293, 2018
|
|
6A5Z
 
 | | Crystal structure of human FXR/RXR-LBD heterodimer bound to HNC180 and 9cRA and SRC1 | | Descriptor: | (9cis)-retinoic acid, 2-[(1R,5S)-9-[[3-[2,6-bis(chloranyl)phenyl]-5-cyclopropyl-1,2-oxazol-4-yl]methoxy]-3-azabicyclo[3.3.1]nonan-3-yl]-1,3-benzothiazole-6-carboxylic acid, Bile acid receptor, ... | | Authors: | Wang, N, Liu, J. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Ligand binding and heterodimerization with retinoid X receptor alpha (RXR alpha ) induce farnesoid X receptor (FXR) conformational changes affecting coactivator binding
J. Biol. Chem., 293, 2018
|
|
6KBU
 
 | | Crystal structure of yedK | | Descriptor: | GLYCEROL, SOS response-associated protein | | Authors: | Wang, N, Bao, H, Huang, H. | | Deposit date: | 2019-06-26 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis of abasic site sensing in single-stranded DNA by the SRAP domain of E. coli yedK.
Nucleic Acids Res., 47, 2019
|
|
6KIJ
 
 | | Crystal structure of yedK with ssDNA containing an abasic site | | Descriptor: | DNA (5'-D(*GP*AP*TP*TP*CP*GP*TP*CP*G)-3'), GLYCEROL, PENTANE-3,4-DIOL-5-PHOSPHATE, ... | | Authors: | Wang, N, Bao, H, Huang, H. | | Deposit date: | 2019-07-18 | | Release date: | 2019-08-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Molecular basis of abasic site sensing in single-stranded DNA by the SRAP domain of E. coli yedK.
Nucleic Acids Res., 47, 2019
|
|
6KBZ
 
 | |
6KBX
 
 | |
6KBS
 
 | | Crystal structure of yedK in complex with ssDNA | | Descriptor: | DNA (5'-D(*CP*GP*GP*TP*CP*GP*AP*TP*TP*C)-3'), SOS response-associated protein | | Authors: | Wang, N, Bao, H, Huang, H. | | Deposit date: | 2019-06-26 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Molecular basis of abasic site sensing in single-stranded DNA by the SRAP domain of E. coli yedK.
Nucleic Acids Res., 47, 2019
|
|
6KCQ
 
 | |
8XKS
 
 | | The cryo-EM structure of Orf2971-FtsHi motor complex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 4Fe-4S ferredoxin-type domain-containing protein, ... | | Authors: | Wang, N, Li, M. | | Deposit date: | 2023-12-24 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Architecture of the ATP-driven motor for protein import into chloroplasts.
Mol Plant, 17, 2024
|
|
8WX0
 
 | | PNPase of M.tuberculosis with its RNA substrate | | Descriptor: | Bifunctional guanosine pentaphosphate synthetase/polyribonucleotide nucleotidyltransferase, RNA (24-mer) | | Authors: | Wang, N, Sheng, Y.N. | | Deposit date: | 2023-10-27 | | Release date: | 2024-07-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of Mycobacterium tuberculosis polynucleotide phosphorylase suggest a potential mechanism for its RNA substrate degradation.
Arch.Biochem.Biophys., 754, 2024
|
|
8WXF
 
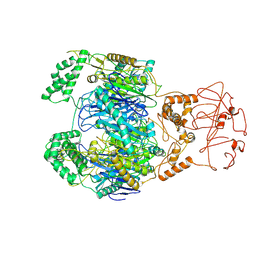 | | PNPase of Mycobacterium tuberculosis | | Descriptor: | Bifunctional guanosine pentaphosphate synthetase/polyribonucleotide nucleotidyltransferase | | Authors: | Wang, N, Sheng, Y.N, Liu, Y.T. | | Deposit date: | 2023-10-29 | | Release date: | 2024-07-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of Mycobacterium tuberculosis polynucleotide phosphorylase suggest a potential mechanism for its RNA substrate degradation.
Arch.Biochem.Biophys., 754, 2024
|
|
7DA5
 
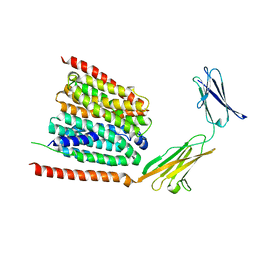 | | Cryo-EM structure of the human MCT1 D309N mutant in complex with Basigin-2 in the inward-open conformation. | | Descriptor: | Basigin, Monocarboxylate transporter 1 | | Authors: | Wang, N, Jiang, X, Zhang, S, Zhu, A, Yuan, Y, Lei, J, Yan, C. | | Deposit date: | 2020-10-14 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of human monocarboxylate transporter 1 inhibition by anti-cancer drug candidates.
Cell, 184, 2021
|
|
7CKR
 
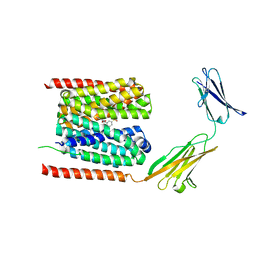 | | Cryo-EM structure of the human MCT1/Basigin-2 complex in the presence of anti-cancer drug candidate BAY-8002 in the outward-open conformation. | | Descriptor: | 2-[[2-chloranyl-5-(phenylsulfonyl)phenyl]carbonylamino]benzoic acid, Basigin, Monocarboxylate transporter 1 | | Authors: | Wang, N, Jiang, X, Zhang, S, Zhu, A, Yuan, Y, Lei, J, Yan, C. | | Deposit date: | 2020-07-18 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of human monocarboxylate transporter 1 inhibition by anti-cancer drug candidates.
Cell, 184, 2021
|
|
7CKO
 
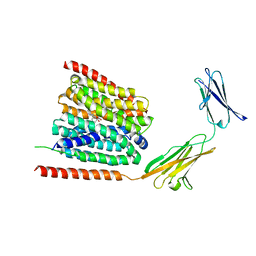 | | Cryo-EM structure of the human MCT1/Basigin-2 complex in the presence of anti-cancer drug candidate 7ACC2 in the inward-open conformation | | Descriptor: | 7-[methyl-(phenylmethyl)amino]-2-oxidanylidene-chromene-3-carboxylic acid, Basigin, Monocarboxylate transporter 1 | | Authors: | Wang, N, Jiang, X, Zhang, S, Zhu, A, Yuan, Y, Lei, J, Yan, C. | | Deposit date: | 2020-07-18 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis of human monocarboxylate transporter 1 inhibition by anti-cancer drug candidates.
Cell, 184, 2021
|
|
6M6I
 
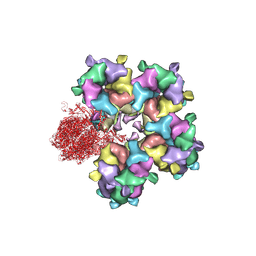 | | Structure of HSV2 B-capsid portal vertex | | Descriptor: | Coiled coils chain 1, Coiled coils chain 2, Major capsid protein, ... | | Authors: | Wang, X.X, Wang, N. | | Deposit date: | 2020-03-14 | | Release date: | 2021-03-10 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Structures of the portal vertex reveal essential protein-protein interactions for Herpesvirus assembly and maturation.
Protein Cell, 11, 2020
|
|
6M6H
 
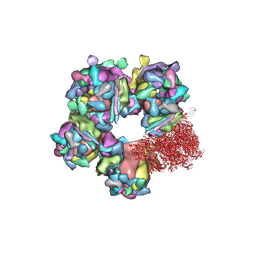 | | Structure of HSV2 C-capsid portal vertex | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | Authors: | Wang, X.X, Wang, N. | | Deposit date: | 2020-03-14 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structures of the portal vertex reveal essential protein-protein interactions for Herpesvirus assembly and maturation.
Protein Cell, 11, 2020
|
|
6M6G
 
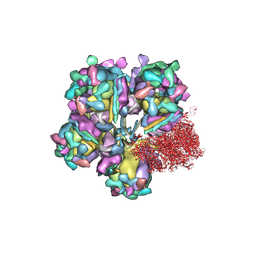 | | Structure of HSV2 viron capsid portal vertex | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Coiled coils, ... | | Authors: | Wang, X.X, Wang, N. | | Deposit date: | 2020-03-14 | | Release date: | 2021-03-24 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (5.39 Å) | | Cite: | Structures of the portal vertex reveal essential protein-protein interactions for Herpesvirus assembly and maturation.
Protein Cell, 11, 2020
|
|
6C6Z
 
 | |
5VZR
 
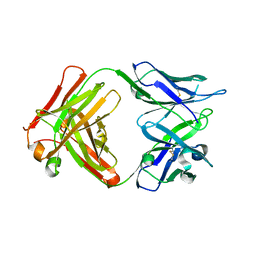 | |
5VYH
 
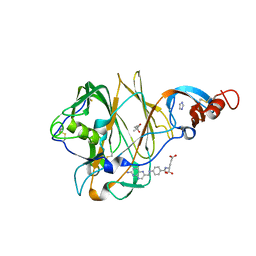 | | Crystal Structure of MERS-CoV S1 N-terminal Domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, FOLIC ACID, ... | | Authors: | Wang, N, Wrapp, D, Pallesen, J, Ward, A.B, McLellan, J.S. | | Deposit date: | 2017-05-25 | | Release date: | 2017-08-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8XA3
 
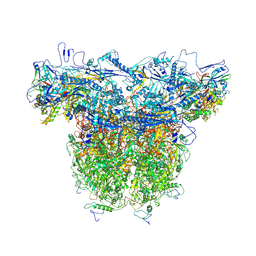 | | C-hexon capsomer of the VZV B-Capsid | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Tri1, ... | | Authors: | Wang, N, Cao, L, Wang, X. | | Deposit date: | 2023-12-01 | | Release date: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Insights into varicella-zoster virus assembly from the B- and C-capsid at near-atomic resolution structures
Hlife, 2, 2024
|
|
7CWM
 
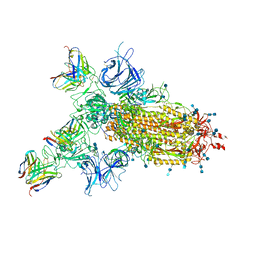 | |
7CWN
 
 | | P17-H014 Fab cocktail in complex with SARS-CoV-2 spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, N, Wang, X. | | Deposit date: | 2020-08-29 | | Release date: | 2020-12-16 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Rational development of a human antibody cocktail that deploys multiple functions to confer Pan-SARS-CoVs protection.
Cell Res., 31, 2021
|
|
