3LXO
 
 | | The crystal structure of ribonuclease A in complex with thymidine-3'-monophosphate | | Descriptor: | Ribonuclease pancreatic, THYMIDINE-3'-PHOSPHATE | | Authors: | Doucet, N, Jayasundera, T.B, Simonovic, M, Loria, J.P. | | Deposit date: | 2010-02-25 | | Release date: | 2010-04-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | The crystal structure of ribonuclease A in complex with thymidine-3'-monophosphate provides further insight into ligand binding.
Proteins, 78, 2010
|
|
7MDL
 
 | | High-resolution crystal structure of human SepSecS-tRNASec complex | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, CITRATE ANION, O-phosphoseryl-tRNA(Sec) selenium transferase, ... | | Authors: | Puppala, A, French, R.L, Simonovic, M. | | Deposit date: | 2021-04-05 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural basis for the tRNA-dependent activation of the terminal complex of selenocysteine synthesis in humans.
Nucleic Acids Res., 2023
|
|
4WYN
 
 | |
3HL2
 
 | | The crystal structure of the human SepSecS-tRNASec complex | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, Monothiophosphate, O-phosphoseryl-tRNA(Sec) selenium transferase, ... | | Authors: | Palioura, S, Steitz, T.A, Soll, D, Simonovic, M. | | Deposit date: | 2009-05-26 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The human SepSecS-tRNASec complex reveals the mechanism of selenocysteine formation.
Science, 325, 2009
|
|
4YE6
 
 | |
4YE9
 
 | |
4WYZ
 
 | | The crystal structure of the A109G mutant of RNase A in complex with 3'UMP | | Descriptor: | 3'-URIDINEMONOPHOSPHATE, Ribonuclease pancreatic | | Authors: | French, R.L, Gagne, D, Doucet, N, Simonovic, M. | | Deposit date: | 2014-11-18 | | Release date: | 2015-11-18 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Perturbation of the Conformational Dynamics of an Active-Site Loop Alters Enzyme Activity.
Structure, 23, 2015
|
|
4WYP
 
 | | The crystal structure of the A109G mutant of RNase A in complex with 5'AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Ribonuclease pancreatic | | Authors: | French, R.L, Gagne, D, Doucet, N, Simonovic, M. | | Deposit date: | 2014-11-17 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Perturbation of the Conformational Dynamics of an Active-Site Loop Alters Enzyme Activity.
Structure, 23, 2015
|
|
4YE8
 
 | |
4ZDP
 
 | |
4YYE
 
 | |
7L1T
 
 | | Crystal structure of human holo SepSecS | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, O-phosphoseryl-tRNA(Sec) selenium transferase, PHOSPHATE ION | | Authors: | Puppala, A, Castillo Suchkou, J, Simonovic, M. | | Deposit date: | 2020-12-15 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for the tRNA-dependent activation of the terminal complex of selenocysteine synthesis in humans.
Nucleic Acids Res., 2023
|
|
1OO8
 
 | |
1OPH
 
 | | NON-COVALENT COMPLEX BETWEEN ALPHA-1-PI-PITTSBURGH AND S195A TRYPSIN | | Descriptor: | Alpha-1-antitrypsin precursor, Trypsinogen, cationic precursor | | Authors: | Dementiev, A, Simonovic, M, Volz, K, Gettins, P.G. | | Deposit date: | 2003-03-05 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Canonical inhibitor-like interactions explain reactivity of alpha1-proteinase inhibitor Pittsburgh and antithrombin with proteinases
J.Biol.Chem., 278, 2003
|
|
1GBQ
 
 | | SOLUTION NMR STRUCTURE OF THE GRB2 N-TERMINAL SH3 DOMAIN COMPLEXED WITH A TEN-RESIDUE PEPTIDE DERIVED FROM SOS DIRECT REFINEMENT AGAINST NOES, J-COUPLINGS, AND 1H AND 13C CHEMICAL SHIFTS, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | GRB2, SOS-1 | | Authors: | Wittekind, M, Mapelli, C, Lee, V, Goldfarb, V, Friedrichs, M.S, Meyers, C.A, Mueller, L. | | Deposit date: | 1996-12-23 | | Release date: | 1997-09-04 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Grb2 N-terminal SH3 domain complexed with a ten-residue peptide derived from SOS: direct refinement against NOEs, J-couplings and 1H and 13C chemical shifts.
J.Mol.Biol., 267, 1997
|
|
1EHC
 
 | | STRUCTURE OF SIGNAL TRANSDUCTION PROTEIN CHEY | | Descriptor: | CHEY, SULFATE ION | | Authors: | Jiang, M, Bourret, R, Simon, M, Volz, K. | | Deposit date: | 1996-03-05 | | Release date: | 1997-05-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Uncoupled phosphorylation and activation in bacterial chemotaxis. The 2.3 A structure of an aspartate to lysine mutant at position 13 of CheY.
J.Biol.Chem., 272, 1997
|
|
8T9H
 
 | |
3C1B
 
 | | The effect of H3 K79 dimethylation and H4 K20 trimethylation on nucleosome and chromatin structure | | Descriptor: | Histone 2, H2bf, Histone H2A type 1, ... | | Authors: | Lu, X, Simon, M, Chodaparambil, J, Hansen, J, Shokat, K, Luger, K. | | Deposit date: | 2008-01-22 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The effect of H3K79 dimethylation and H4K20 trimethylation on nucleosome and chromatin structure.
Nat.Struct.Mol.Biol., 15, 2008
|
|
3C1C
 
 | | The effect of H3 K79 dimethylation and H4 K20 trimethylation on nucleosome and chromatin structure | | Descriptor: | Histone 2, H2bf, Histone H2A type 1, ... | | Authors: | Lu, X, Simon, M, Chodaparambil, J, Hansen, J, Shokat, K, Luger, K. | | Deposit date: | 2008-01-22 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The effect of H3K79 dimethylation and H4K20 trimethylation on nucleosome and chromatin structure.
Nat.Struct.Mol.Biol., 15, 2008
|
|
5A0E
 
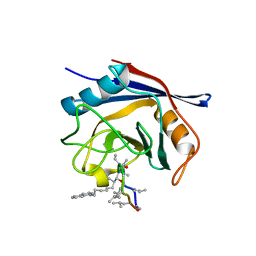 | | Crystal structure of cyclophilin D in complex with CsA analogue, JW47. | | Descriptor: | JW47, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE F, MITOCHONDRIAL | | Authors: | Warne, J, Pryce, G, Hill, J, Shi, X, Lenneras, F, Puentes, F, Kip, M, Hilditch, L, Walker, P, Simone, M, Chan, A.W.E, Towers, G, Coker, A.R, Duchen, M, Szabadkai, G, Baker, D, Selwood, D.L. | | Deposit date: | 2015-04-19 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Selective Inhibition of the Mitochondrial Permeability Transition Pore Protects Against Neuro-Degeneration in Experimental Multiple Sclerosis.
J.Biol.Chem., 291, 2016
|
|
3GBQ
 
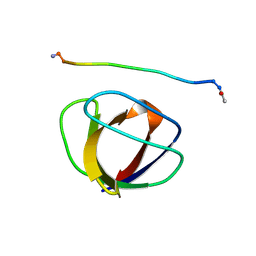 | | SOLUTION NMR STRUCTURE OF THE GRB2 N-TERMINAL SH3 DOMAIN COMPLEXED WITH A TEN-RESIDUE PEPTIDE DERIVED FROM SOS DIRECT REFINEMENT AGAINST NOES, J-COUPLINGS, AND 1H AND 13C CHEMICAL SHIFTS, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | GRB2, SOS-1 | | Authors: | Wittekind, M, Mapelli, C, Lee, V, Goldfarb, V, Friedrichs, M.S, Meyers, C.A, Mueller, L. | | Deposit date: | 1996-12-23 | | Release date: | 1997-09-04 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Grb2 N-terminal SH3 domain complexed with a ten-residue peptide derived from SOS: direct refinement against NOEs, J-couplings and 1H and 13C chemical shifts.
J.Mol.Biol., 267, 1997
|
|
4GBQ
 
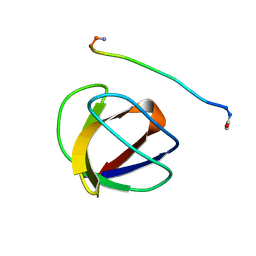 | | SOLUTION NMR STRUCTURE OF THE GRB2 N-TERMINAL SH3 DOMAIN COMPLEXED WITH A TEN-RESIDUE PEPTIDE DERIVED FROM SOS DIRECT REFINEMENT AGAINST NOES, J-COUPLINGS, AND 1H AND 13C CHEMICAL SHIFTS, 15 STRUCTURES | | Descriptor: | GRB2, SOS-1 | | Authors: | Wittekind, M, Mapelli, C, Lee, V, Goldfarb, V, Friedrichs, M.S, Meyers, C.A, Mueller, L. | | Deposit date: | 1996-12-23 | | Release date: | 1997-09-04 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Grb2 N-terminal SH3 domain complexed with a ten-residue peptide derived from SOS: direct refinement against NOEs, J-couplings and 1H and 13C chemical shifts.
J.Mol.Biol., 267, 1997
|
|
1GBR
 
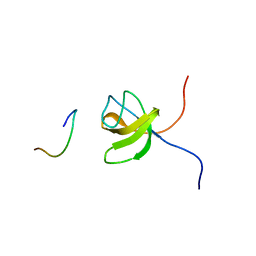 | | ORIENTATION OF PEPTIDE FRAGMENTS FROM SOS PROTEINS BOUND TO THE N-TERMINAL SH3 DOMAIN OF GRB2 DETERMINED BY NMR SPECTROSCOPY | | Descriptor: | GROWTH FACTOR RECEPTOR-BOUND PROTEIN 2, SOS-A PEPTIDE | | Authors: | Wittekind, M, Mapelli, C, Farmer, B.T, Suen, K.-L, Goldfarb, V, Tsao, J, Lavoie, T, Barbacid, M, Meyers, C.A, Mueller, L. | | Deposit date: | 1994-08-12 | | Release date: | 1995-01-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Orientation of peptide fragments from Sos proteins bound to the N-terminal SH3 domain of Grb2 determined by NMR spectroscopy.
Biochemistry, 33, 1994
|
|
2GBQ
 
 | | SOLUTION NMR STRUCTURE OF THE GRB2 N-TERMINAL SH3 DOMAIN COMPLEXED WITH A TEN-RESIDUE PEPTIDE DERIVED FROM SOS DIRECT REFINEMENT AGAINST NOES, J-COUPLINGS, AND 1H AND 13C CHEMICAL SHIFTS, 15 STRUCTURES | | Descriptor: | GRB2, SOS-1 | | Authors: | Wittekind, M, Mapelli, C, Lee, V, Goldfarb, V, Friedrichs, M.S, Meyers, C.A, Mueller, L. | | Deposit date: | 1996-12-23 | | Release date: | 1997-09-04 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Grb2 N-terminal SH3 domain complexed with a ten-residue peptide derived from SOS: direct refinement against NOEs, J-couplings and 1H and 13C chemical shifts.
J.Mol.Biol., 267, 1997
|
|
1HCR
 
 | |
