2GGE
 
 | | Crystal Structure of Mandelate Racemase/Muconate Lactonizing Enzyme from Bacillus Subtilis complexed with MG++ at 1.8 A | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, yitF | | Authors: | Malashkevich, V.N, Sauder, J.M, Schwinn, K.D, Emtage, S, Thompson, D.A, Rutter, M.E, Dickey, M, Groshong, C, Bain, K.T, Adams, J.M, Reyes, C, Rooney, I, Powell, A, Boice, A, Gheyi, T, Ozyurt, S, Atwell, S, Wasserman, S.R, Burley, S.K, Sali, A, Babbitt, P, Pieper, U, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-03-23 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal Structure of Mandelate Racemase/Muconate Lactonizing Enzyme from Bacillus Subtilis complexed with MG++ at 1.8 A
To be Published
|
|
1FI4
 
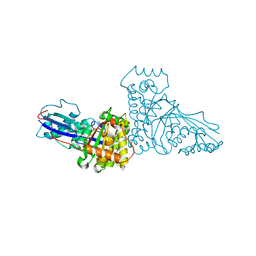 | | THE X-RAY CRYSTAL STRUCTURE OF MEVALONATE 5-DIPHOSPHATE DECARBOXYLASE AT 2.3 ANGSTROM RESOLUTION. | | Descriptor: | MEVALONATE 5-DIPHOSPHATE DECARBOXYLASE | | Authors: | Bonanno, J.B, Edo, C, Eswar, N, Pieper, U, Romanowski, M.J, Ilyin, V, Gerchman, S.E, Kycia, H, Studier, F.W, Sali, A, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2000-08-03 | | Release date: | 2001-03-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural genomics of enzymes involved in sterol/isoprenoid biosynthesis.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1G61
 
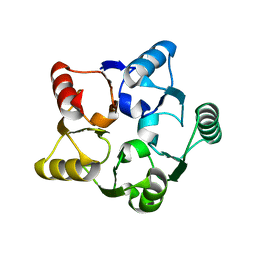 | | CRYSTAL STRUCTURE OF M.JANNASCHII EIF6 | | Descriptor: | TRANSLATION INITIATION FACTOR 6 | | Authors: | Groft, C.M, Beckmann, R, Sali, A, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2000-11-02 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structures of ribosome anti-association factor IF6.
Nat.Struct.Biol., 7, 2000
|
|
1G62
 
 | | CRYSTAL STRUCTURE OF S.CEREVISIAE EIF6 | | Descriptor: | RIBOSOME ANTI-ASSOCIATION FACTOR EIF6 | | Authors: | Groft, C.M, Beckmann, R, Sali, A, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2000-11-02 | | Release date: | 2000-11-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of ribosome anti-association factor IF6.
Nat.Struct.Biol., 7, 2000
|
|
4TQG
 
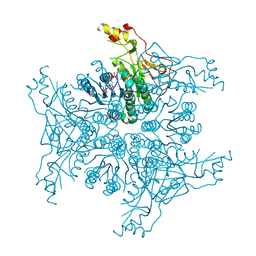 | | Crystal structure of Megavirus UDP-GlcNAc 4,6-dehydratase, 5-epimerase Mg534 | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative dTDP-d-glucose 4 6-dehydratase | | Authors: | Jeudy, S, Piacente, F, De Castro, C, Molinaro, A, Salis, A, Damonte, G, Bernardi, C, Tonetti, M, Claverie, J.M, Abergel, C. | | Deposit date: | 2014-06-11 | | Release date: | 2014-06-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Giant Virus Megavirus chilensis Encodes the Biosynthetic Pathway for Uncommon Acetamido Sugars.
J.Biol.Chem., 289, 2014
|
|
4AEC
 
 | | Crystal Structure of the Arabidopsis thaliana O-Acetyl-Serine-(Thiol)- Lyase C | | Descriptor: | ACETATE ION, CYSTEINE SYNTHASE, MITOCHONDRIAL, ... | | Authors: | Feldman-Salit, A, Wirtz, M, Lenherr, E.D, Throm, C, Hothorn, M, Scheffzek, K, Hell, R, Wade, R.C. | | Deposit date: | 2012-01-09 | | Release date: | 2012-02-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Allosterically Gated Enzyme Dynamics in the Cysteine Synthase Complex Regulate Cysteine Biosynthesis in Arabidopsis Thaliana.
Structure, 20, 2012
|
|
6D35
 
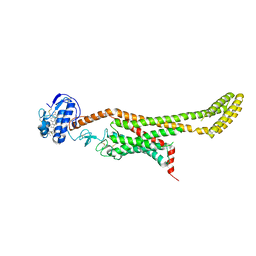 | | Crystal structure of Xenopus Smoothened in complex with cholesterol | | Descriptor: | CHOLESTEROL, Smoothened,Soluble cytochrome b562,Smoothened | | Authors: | Huang, P, Zheng, S, Kim, Y, Kruse, A.C, Salic, A. | | Deposit date: | 2018-04-14 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural Basis of Smoothened Activation in Hedgehog Signaling.
Cell, 174, 2018
|
|
6D32
 
 | | Crystal structure of Xenopus Smoothened in complex with cyclopamine | | Descriptor: | Cyclopamine, Smoothened,Soluble cytochrome b562,Smoothened | | Authors: | Huang, P, Zheng, S, Kim, Y, Kruse, A.C, Salic, A. | | Deposit date: | 2018-04-14 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.751 Å) | | Cite: | Structural Basis of Smoothened Activation in Hedgehog Signaling.
Cell, 174, 2018
|
|
5KZV
 
 | | Crystal structure of the xenopus Smoothened cysteine-rich domain (CRD) in complex with 20(S)-hydroxycholesterol | | Descriptor: | (3alpha,8alpha)-cholest-5-ene-3,20-diol, Smoothened | | Authors: | Huang, P, Kim, Y, Salic, A. | | Deposit date: | 2016-07-25 | | Release date: | 2016-08-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.616 Å) | | Cite: | Cellular Cholesterol Directly Activates Smoothened in Hedgehog Signaling.
Cell, 166, 2016
|
|
5KZY
 
 | |
5KZZ
 
 | | Crystal structure of the xenopus Smoothened cysteine-rich domain (CRD) in its apo-form | | Descriptor: | ACETATE ION, GLYCEROL, Smoothened, ... | | Authors: | Huang, P, Kim, Y, Salic, A. | | Deposit date: | 2016-07-25 | | Release date: | 2016-08-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.332 Å) | | Cite: | Cellular Cholesterol Directly Activates Smoothened in Hedgehog Signaling.
Cell, 166, 2016
|
|
7RHR
 
 | | Cryo-EM structure of Xenopus Patched-1 in nanodisc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Huang, P, Lian, T, Jiang, J, Salic, A. | | Deposit date: | 2021-07-18 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for catalyzed assembly of the Sonic hedgehog-Patched1 signaling complex.
Dev.Cell, 57, 2022
|
|
7RHQ
 
 | | Cryo-EM structure of Xenopus Patched-1 in complex with GAS1 and Sonic Hedgehog | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Huang, P, Lian, T, Wierbowski, B, Garcia-Linares, S, Jiang, J, Salic, A. | | Deposit date: | 2021-07-18 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Structural basis for catalyzed assembly of the Sonic hedgehog-Patched1 signaling complex.
Dev.Cell, 57, 2022
|
|
2Q5E
 
 | | Crystal structure of human carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 2 | | Descriptor: | Carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 2, MAGNESIUM ION | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Lau, C, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-05-31 | | Release date: | 2007-06-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
4XX9
 
 | |
6N1Z
 
 | | Importin-9 bound to H2A-H2B | | Descriptor: | Histone H2A, Histone H2B 1.1, Importin-9 | | Authors: | Tomchick, D.R, Chook, Y.M, Padavannil, A. | | Deposit date: | 2018-11-12 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Importin-9 wraps around the H2A-H2B core to act as nuclear importer and histone chaperone.
Elife, 8, 2019
|
|
6URI
 
 | |
8G0I
 
 | | High Affinity nanobodies against GFP | | Descriptor: | CHLORIDE ION, Green fluorescent protein, LaG24 Nanobody, ... | | Authors: | Ketaren, N.E, Rout, M.P, Almo, S. | | Deposit date: | 2023-01-31 | | Release date: | 2024-03-20 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unique Binding and Stabilization Mechanisms Employed By and Engineered Into Nanobodies
Biorxiv, 2023
|
|
5LN3
 
 | | The human 26S Proteasome at 6.8 Ang. | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Schweitzer, A, Beck, F, Sakata, E, Unverdorben, P. | | Deposit date: | 2016-08-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Molecular Details Underlying Dynamic Structures and Regulation of the Human 26S Proteasome.
Mol. Cell Proteomics, 16, 2017
|
|
2FH7
 
 | | Crystal structure of the phosphatase domains of human PTP SIGMA | | Descriptor: | Receptor-type tyrosine-protein phosphatase S | | Authors: | Alvarado, J, Udupi, R, Smith, D, Koss, J, Wasserman, S.R, Ozyurt, S, Atwell, S, Powell, A, Kearins, M.C, Rooney, I, Maletic, M, Bain, K.T, Freeman, J.C, Russell, M, Thompson, D.A, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-12-23 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
7KDT
 
 | |
5TVZ
 
 | | Solution NMR structure of Saccharomyces cerevisiae Pom152 Ig-like repeat, residues 718-820 | | Descriptor: | Nucleoporin POM152 | | Authors: | Dutta, K, Sampathkumar, P, Cowburn, D, Almo, S.C, Rout, M.P, Fernandez-Martinez, J. | | Deposit date: | 2016-11-10 | | Release date: | 2017-02-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular Architecture of the Major Membrane Ring Component of the Nuclear Pore Complex.
Structure, 25, 2017
|
|
3TXM
 
 | |
3TXN
 
 | |
2ER7
 
 | | X-RAY ANALYSES OF ASPARTIC PROTEINASES.III. THREE-DIMENSIONAL STRUCTURE OF ENDOTHIAPEPSIN COMPLEXED WITH A TRANSITION-STATE ISOSTERE INHIBITOR OF RENIN AT 1.6 ANGSTROMS RESOLUTION | | Descriptor: | ENDOTHIAPEPSIN, SULFATE ION, TRANSITION-STATE ISOSTERE INHIBITOR OF RENIN | | Authors: | Veerapandian, B, Cooper, J.B, Szelke, M, Blundell, T.L. | | Deposit date: | 1990-11-12 | | Release date: | 1991-01-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray analyses of aspartic proteinases. III Three-dimensional structure of endothiapepsin complexed with a transition-state isostere inhibitor of renin at 1.6 A resolution.
J.Mol.Biol., 216, 1990
|
|
