7O6W
 
 | | Crystal structure of (the) VEL1 VEL polymerising domain (I664D mutant) | | Descriptor: | PHOSPHATE ION, VIN3-like protein 2 | | Authors: | Fiedler, M, Franco-Echevarria, E, Dean, C, Bienz, M. | | Deposit date: | 2021-04-12 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Head-to-tail polymerization by VEL proteins underpins cold-induced Polycomb silencing in flowering control.
Cell Rep, 41, 2022
|
|
7OQV
 
 | |
2BTG
 
 | |
2BTH
 
 | |
6S9T
 
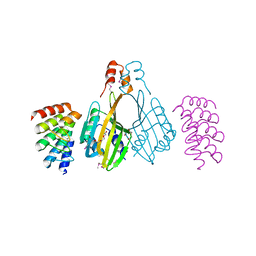 | | Dimerization domain of Xenopus laevis LDB1 in complex with darpin 3 | | Descriptor: | Darpin 3, LIM domain-binding protein 1, TETRAETHYLENE GLYCOL | | Authors: | Renko, M, Schaefer, J.V, Pluckthun, A, Bienz, M. | | Deposit date: | 2019-07-15 | | Release date: | 2019-10-09 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Rotational symmetry of the structured Chip/LDB-SSDP core module of the Wnt enhanceosome.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6S9S
 
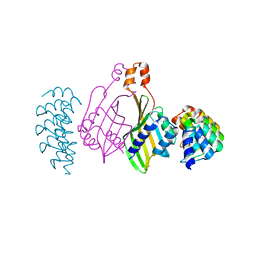 | | Dimerization domain of Xenopus laevis LDB1 in complex with darpin 10 | | Descriptor: | Darpin 10, LIM domain-binding protein 1 | | Authors: | Renko, M, Schaefer, J.V, Pluckthun, A, Bienz, M. | | Deposit date: | 2019-07-15 | | Release date: | 2019-10-09 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rotational symmetry of the structured Chip/LDB-SSDP core module of the Wnt enhanceosome.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6S9R
 
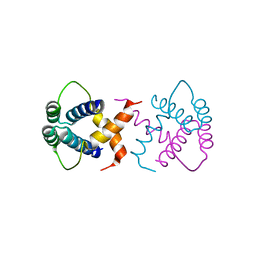 | | Crystal structure of SSDP from D. melanogaster | | Descriptor: | Sequence-specific single-stranded DNA-binding protein, isoform A | | Authors: | Renko, M, Bienz, M. | | Deposit date: | 2019-07-15 | | Release date: | 2019-10-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Rotational symmetry of the structured Chip/LDB-SSDP core module of the Wnt enhanceosome.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4AGP
 
 | | Structure of the p53 core domain mutant Y220C bound to the stabilizing small molecule PhiKan5176 | | Descriptor: | 2-{[4-(diethylamino)piperidin-1-yl]methyl}-6-iodo-4-(3-phenoxyprop-1-yn-1-yl)phenol, CELLULAR TUMOR ANTIGEN P53, ZINC ION | | Authors: | Joerger, A.C, Wilcken, R, Fersht, A.R, Boeckler, F.M. | | Deposit date: | 2012-01-30 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Halogen-Enriched Fragment Libraries as Leads for Drug Rescue of Mutant P53.
J.Am.Chem.Soc., 134, 2012
|
|
4AGN
 
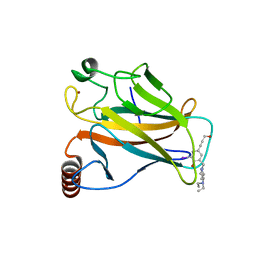 | | Structure of the p53 core domain mutant Y220C bound to the stabilizing small molecule PhiKan5116 | | Descriptor: | 2-{[4-(DIETHYLAMINO)PIPERIDIN-1-YL]METHYL}-4-(3-HYDROXYPROP-1-YN-1-YL)-6-IODOPHENOL, CELLULAR TUMOR ANTIGEN P53, ZINC ION | | Authors: | Joerger, A.C, Wilcken, R, Boeckler, F.M, Fersht, A.R. | | Deposit date: | 2012-01-30 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Halogen-Enriched Fragment Libraries as Leads for Drug Rescue of Mutant P53.
J.Am.Chem.Soc., 134, 2012
|
|
2X0V
 
 | | STRUCTURE OF THE P53 CORE DOMAIN MUTANT Y220C BOUND TO 4-(trifluoromethyl)benzene-1,2-diamine | | Descriptor: | 4-(TRIFLUOROMETHYL)BENZENE-1,2-DIAMINE, CELLULAR TUMOR ANTIGEN P53, ZINC ION | | Authors: | Basse, N, Kaar, J.L, Joerger, A.C, Fersht, A.R. | | Deposit date: | 2009-12-17 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Toward the Rational Design of P53-Stabilizing Drugs: Probing the Surface of the Oncogenic Y220C Mutant.
Chem.Biol., 17, 2010
|
|
2VUK
 
 | |
4AGQ
 
 | | Structure of the p53 core domain mutant Y220C bound to the stabilizing small molecule PhiKan5196 | | Descriptor: | 2-{[4-(diethylamino)piperidin-1-yl]methyl}-6-iodo-4-[3-(phenylamino)prop-1-yn-1-yl]phenol, CELLULAR TUMOR ANTIGEN P53, ZINC ION | | Authors: | Joerger, A.C, Wilcken, R, Boeckler, F.M, Fersht, A.R. | | Deposit date: | 2012-01-30 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Halogen-Enriched Fragment Libraries as Leads for Drug Rescue of Mutant P53.
J.Am.Chem.Soc., 134, 2012
|
|
4AGL
 
 | | Structure of the p53 core domain mutant Y220C bound to the stabilizing small molecule PhiKan784 | | Descriptor: | 2,4-BIS(IODANYL)-6-[[METHYL-(1-METHYLPIPERIDIN-4-YL)AMINO]METHYL]PHENOL, CELLULAR TUMOR ANTIGEN P53, ZINC ION | | Authors: | Joerger, A.C, Wilcken, R, Boeckler, F.M, Fersht, A.R. | | Deposit date: | 2012-01-30 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Halogen-Enriched Fragment Libraries as Leads for Drug Rescue of Mutant P53.
J.Am.Chem.Soc., 134, 2012
|
|
5SUY
 
 | |
4AGM
 
 | | Structure of the p53 core domain mutant Y220C bound to the stabilizing small molecule PhiKan5086 | | Descriptor: | 2-{[4-(DIETHYLAMINO)PIPERIDIN-1-YL]METHYL}-4,6-DIIODOPHENOL, CELLULAR TUMOR ANTIGEN P53, ZINC ION | | Authors: | Joerger, A.C, Wilcken, R, Boeckler, F.M, Fersht, A.R. | | Deposit date: | 2012-01-30 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Halogen-Enriched Fragment Libraries as Leads for Drug Rescue of Mutant P53.
J.Am.Chem.Soc., 134, 2012
|
|
2X0W
 
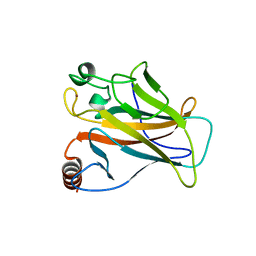 | | STRUCTURE OF THE P53 CORE DOMAIN MUTANT Y220C BOUND TO 5,6-dimethoxy- 2-methylbenzothiazole | | Descriptor: | 5,6-DIMETHOXY-2-METHYL-1,3-BENZOTHIAZOLE, CELLULAR TUMOR ANTIGEN P53, ZINC ION | | Authors: | Kaar, J.L, Basse, N, Joerger, A.C, Fersht, A.R. | | Deposit date: | 2009-12-17 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Toward the Rational Design of P53-Stabilizing Drugs: Probing the Surface of the Oncogenic Y220C Mutant.
Chem.Biol., 17, 2010
|
|
5SUZ
 
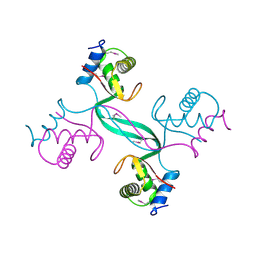 | |
3ZRH
 
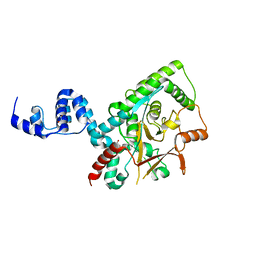 | | Crystal structure of the Lys29, Lys33-linkage-specific TRABID OTU deubiquitinase domain reveals an Ankyrin-repeat ubiquitin binding domain (AnkUBD) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, UBIQUITIN THIOESTERASE ZRANB1 | | Authors: | Licchesi, J.D.F, Akutsu, M, Komander, D. | | Deposit date: | 2011-06-16 | | Release date: | 2011-12-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | An Ankyrin-Repeat Ubiquitin-Binding Domain Determines Trabid'S Specificity for Atypical Ubiquitin Chains.
Nat.Struct.Mol.Biol., 19, 2011
|
|
4AGO
 
 | | Structure of the p53 core domain mutant Y220C bound to the stabilizing small molecule PhiKan5174 | | Descriptor: | CELLULAR TUMOR ANTIGEN P53, TERT-BUTYL [3-(3-{[4-(DIETHYLAMINO)PIPERIDIN-1-YL]METHYL}-4-HYDROXY-5-IODOPHENYL)PROP-2-YN-1-YL]CARBAMATE, ZINC ION | | Authors: | Joerger, A.C, Wilcken, R, Fersht, A.R, Boeckler, F.M. | | Deposit date: | 2012-01-30 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Halogen-Enriched Fragment Libraries as Leads for Drug Rescue of Mutant P53.
J.Am.Chem.Soc., 134, 2012
|
|
2X0U
 
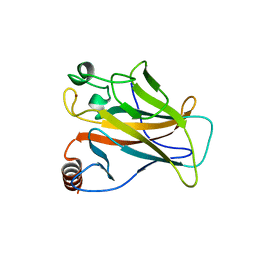 | | STRUCTURE OF THE P53 CORE DOMAIN MUTANT Y220C BOUND TO A 2-amino substituted benzothiazole scaffold | | Descriptor: | 6,7-DIHYDRO[1,4]DIOXINO[2,3-F][1,3]BENZOTHIAZOL-2-AMINE, CELLULAR TUMOR ANTIGEN P53, ZINC ION | | Authors: | Joerger, A.C, Kaar, J.L, Basse, N, Fersht, A.R. | | Deposit date: | 2009-12-17 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Toward the Rational Design of P53-Stabilizing Drugs: Probing the Surface of the Oncogenic Y220C Mutant.
Chem.Biol., 17, 2010
|
|
7O06
 
 | |
7O3B
 
 | |
7O0S
 
 | |
3SL9
 
 | | X-ray structure of Beta catenin in complex with Bcl9 | | Descriptor: | 1,2-ETHANEDIOL, B-cell CLL/lymphoma 9 protein, Catenin beta-1, ... | | Authors: | Gupta, D, Bienz, M. | | Deposit date: | 2011-06-24 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | An intrinsically labile alpha-helix abutting the BCL9-binding site of beta-catenin is required for its inhibition by carnosic acid.
Nat Commun, 3, 2012
|
|
3SLA
 
 | | X-ray structure of first four repeats of human beta-catenin | | Descriptor: | Catenin beta-1, GLYCEROL, SODIUM ION | | Authors: | Gupta, D, Bienz, M. | | Deposit date: | 2011-06-24 | | Release date: | 2012-02-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An intrinsically labile alpha-helix abutting the BCL9-binding site of beta-catenin is required for its inhibition by carnosic acid.
Nat Commun, 3, 2012
|
|
