2BF4
 
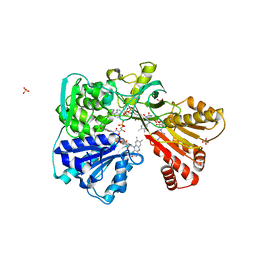 | | A second FMN-binding site in yeast NADPH-cytochrome P450 reductase suggests a novel mechanism of electron transfer by diflavin reductases. | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Podust, L.M, Lepesheva, G.I, Kim, Y, Yermalitskaya, L.V, Yermalitsky, V.N, Lamb, D.C, Kelly, S.L, Waterman, M.R. | | Deposit date: | 2004-12-03 | | Release date: | 2006-01-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Second Fmn-Binding Site in Yeast Nadph-Cytochrome P450 Reductase Suggests a Mechanism of Electron Transfer by Diflavin Reductases.
Structure, 14, 2006
|
|
2BZ9
 
 | |
2BN4
 
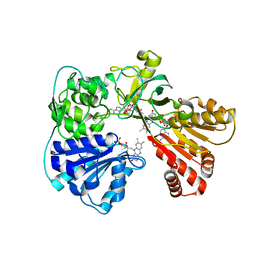 | | A second FMN-binding site in yeast NADPH-cytochrome P450 reductase suggests a novel mechanism of electron transfer by diflavin reductase | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Podust, L.M, Lepesheva, G.I, Kim, Y, Yermalitskaya, L.V, Yermalitsky, V.N, Lamb, D.C, Kelly, S.L, Waterman, M.R. | | Deposit date: | 2005-03-18 | | Release date: | 2006-01-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | A Second Fmn-Binding Site in Yeast Nadph-Cytochrome P450 Reductase Suggests a Mechanism of Electron Transfer by Diflavin Reductases.
Structure, 14, 2006
|
|
4B7S
 
 | |
4BJK
 
 | |
2UVN
 
 | | Crystal structure of econazole-bound CYP130 from Mycobacterium tuberculosis | | Descriptor: | 1-[(2S)-2-[(4-CHLOROBENZYL)OXY]-2-(2,4-DICHLOROPHENYL)ETHYL]-1H-IMIDAZOLE, CYTOCHROME P450 130, FLUORIDE ION, ... | | Authors: | Podust, L.M, Ortiz de Montellano, P.R. | | Deposit date: | 2007-03-12 | | Release date: | 2007-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mycobacterium Tuberculosis Cyp130: Crystal Structure, Biophysical Characterization, and Interactions with Antifungal Azole Drugs
J.Biol.Chem., 283, 2008
|
|
2W0A
 
 | |
2W09
 
 | |
2W0B
 
 | |
2UUQ
 
 | |
4UVR
 
 | | Binding mode, selectivity and potency of N-indolyl-oxopyridinyl-4- amino-propanyl-based inhibitors targeting Trypanosoma cruzi CYP51 | | Descriptor: | Nalpha-{4-[4-(5-chloro-2-methylphenyl)piperazin-1-yl]-2-fluorobenzoyl}-N-pyridin-4-yl-D-tryptophanamide, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-DEMETHYLASE, ... | | Authors: | Vieira, D.F, Choi, J.Y, Calvet, C.M, Gut, J, Kellar, D, Siqueira-Neto, J.L, Johnston, J.B, McKerrow, J.H, Roush, W.R, Podust, L.M. | | Deposit date: | 2014-08-08 | | Release date: | 2014-11-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Binding Mode and Potency of N-Indolyl-Oxopyridinyl-4-Amino-Propanyl-Based Inhibitors Targeting Trypanosoma Cruzi Cyp51
J.Med.Chem., 57, 2014
|
|
7RKT
 
 | |
7RKW
 
 | |
7RKR
 
 | |
4UQH
 
 | | Crystal structure of Trypanosoma cruzi CYP51 bound to the inhibitor (R)-N-(3-(1H-indol-3-yl)-1-oxo-1-(pyridin-4-ylamino)propan-2-yl)-4-(4-(3,4-difluorophenyl)piperazin-1-yl)-2-fluorobenzamide. | | Descriptor: | (R)-N-(3-(1H-indol-3-yl)-1-oxo-1-(pyridin-4-ylamino)propan-2-yl)-4-(4-(3,4-difluorophenyl)piperazin-1-yl)-2-fluorobenzamide, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE, ... | | Authors: | Calvet, C.M, Vieira, D.F, Choi, J.Y, Cameron, M.D, Gut, J, Kellar, D, Siqueira-Neto, J.L, McKerrow, J.H, Roush, W.R, Podust, L.M. | | Deposit date: | 2014-06-23 | | Release date: | 2014-08-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | 4-Aminopyridyl-Based Cyp51 Inhibitors as Anti-Trypanosoma Cruzi Drug Leads with Improved Pharmacokinetic Profile and in Vivo Potency.
J.Med.Chem., 57, 2014
|
|
1JNM
 
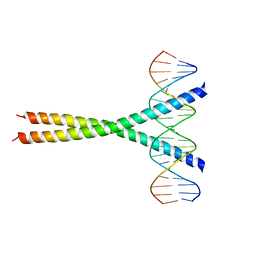 | | Crystal Structure of the Jun/CRE Complex | | Descriptor: | 5'-D(*CP*GP*TP*CP*GP*AP*TP*GP*AP*CP*GP*TP*CP*AP*TP*CP*GP*AP*CP*G)-3', PROTO-ONCOGENE C-JUN | | Authors: | Kim, Y, Podust, L.M. | | Deposit date: | 2001-07-24 | | Release date: | 2003-06-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Jun bZIP homodimer complexed with CRE
To be Published
|
|
6O6E
 
 | | Crystal structure of PltF trapped with PltL using a proline adenosine vinylsulfonamide inhibitor | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-deoxy-5'-({(2S)-2-({2-[(N-{(2R)-4-[(dioxo-lambda~5~-phosphanyl)oxy]-2-hydroxy-3,3-dimethylbutanoyl}-beta-alanyl)amino]ethyl}sulfanyl)-2-[(2S)-pyrrolidin-2-yl]ethanesulfonyl}amino)adenosine, FORMIC ACID, ... | | Authors: | Corpuz, J.C, Podust, L.M. | | Deposit date: | 2019-03-06 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Dynamic visualization of type II peptidyl carrier protein recognition in pyoluteorin biosynthesis.
Rsc Chem Biol, 1, 2020
|
|
6Q2C
 
 | | Domain-swapped dimer of Acanthamoeba castellanii CYP51 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Obtusifoliol 14alphademethylase, ... | | Authors: | Sharma, V, Podust, L.M. | | Deposit date: | 2019-08-07 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Domain-Swap Dimerization of Acanthamoeba castellanii CYP51 and a Unique Mechanism of Inactivation by Isavuconazole.
Mol.Pharmacol., 98, 2020
|
|
6UW2
 
 | | Clotrimazole bound complex of Acanthamoeba castellanii CYP51 | | Descriptor: | 1-[(2-CHLOROPHENYL)(DIPHENYL)METHYL]-1H-IMIDAZOLE, GUANIDINE, Obtusifoliol 14alphademethylase, ... | | Authors: | Sharma, V, Podust, L.M. | | Deposit date: | 2019-11-04 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Domain-Swap Dimerization of Acanthamoeba castellanii CYP51 and a Unique Mechanism of Inactivation by Isavuconazole.
Mol.Pharmacol., 98, 2020
|
|
6UX0
 
 | | Isavuconazole bound complex of Acanthamoeba castellanii CYP51 | | Descriptor: | 4-{2-[(2R,3R)-3-(2,5-difluorophenyl)-3-hydroxy-4-(1H-1,2,4-triazol-1-yl)butan-2-yl]-1,3-thiazol-4-yl}benzonitrile, FE (III) ION, Obtusifoliol 14alphademethylase, ... | | Authors: | Sharma, V, Podust, L.M. | | Deposit date: | 2019-11-06 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Domain-Swap Dimerization of Acanthamoeba castellanii CYP51 and a Unique Mechanism of Inactivation by Isavuconazole.
Mol.Pharmacol., 98, 2020
|
|
5FOI
 
 | | Crystal structure of mycinamicin VIII C21 methyl hydroxylase MycCI from Micromonospora griseorubida bound to mycinamicin VIII | | Descriptor: | GLYCEROL, MYCINAMICIN VIII C21 METHYL HYDROXYLASE, Mycinamicin VIII, ... | | Authors: | Demars, M, Sheng, F, Podust, L.M, Sherman, D.H. | | Deposit date: | 2015-11-22 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Biochemical and Structural Characterization of Mycci, a Versatile P450 Biocatalyst from the Mycinamicin Biosynthetic Pathway.
Acs Chem.Biol., 11, 2016
|
|
6XA2
 
 | | Structure of the tirandamycin C-bound P450 monooxygenase TamI | | Descriptor: | (3E)-3-{(2E,4E,6R)-1-hydroxy-4-methyl-6-[(1R,3R,4S,5R)-1,4,8-trimethyl-2,9-dioxabicyclo[3.3.1]non-7-en-3-yl]hepta-2,4-dien-1-ylidene}-2H-pyrrole-2,4(3H)-dione, PROTOPORPHYRIN IX CONTAINING FE, TamI | | Authors: | Newmister, S.A, Srivastava, K.R, Espinoza, R.V, Haatveit, K.C, Khatri, Y, Martini, R.M, Garcia-Borras, M, Podust, L.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2020-06-03 | | Release date: | 2021-06-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Molecular Basis of Iterative C─H Oxidation by TamI, a Multifunctional P450 monooxygenase from the Tirandamycin Biosynthetic Pathway.
Acs Catalysis, 10, 2020
|
|
6XA3
 
 | | Structure of the ligand free P450 monooxygenase TamI | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, TamI | | Authors: | Newmister, S.A, Srivastava, K.R, Espinoza, R.V, Haatveit, K.C, Khatri, Y, Martini, R.M, Garcia-Borras, M, Podust, L.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2020-06-03 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Molecular Basis of Iterative C─H Oxidation by TamI, a Multifunctional P450 monooxygenase from the Tirandamycin Biosynthetic Pathway.
Acs Catalysis, 10, 2020
|
|
3ZBY
 
 | | Ligand-free structure of CYP142 from Mycobacterium smegmatis | | Descriptor: | 1,2-ETHANEDIOL, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), P450 HEME-THIOLATE PROTEIN, ... | | Authors: | Garcia-Fernandez, E, Frank, D.J, Galan, B, Kells, P.M, Podust, L.M, Garcia, J.L, Ortiz de Montellano, P.R. | | Deposit date: | 2012-11-13 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | A Highly Conserved Mycobacterial Cholesterol Catabolic Pathway.
Environ.Microbiol., 15, 2013
|
|
3ZSN
 
 | | Structure of the mixed-function P450 MycG F286A mutant in complex with mycinamicin IV | | Descriptor: | BENZAMIDINE, GLYCEROL, MYCINAMICIN IV, ... | | Authors: | Li, S, Kells, P.M, Rutaganira, F.U, Anzai, Y, Kato, F, Sherman, D.H, Podust, L.M. | | Deposit date: | 2011-06-29 | | Release date: | 2012-05-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substrate Recognition by the Multifunctional Cytochrome P450 Mycg in Mycinamicin Hydroxylation and Epoxidation Reactions.
J.Biol.Chem., 287, 2012
|
|
