6YV2
 
 | | STRUCTURE OF THE WNT DEACYLASE NOTUM IN COMPLEX WITH A PYRROLIDINE-3-CARBOXYLIC ACID FRAGMENT 598 | | Descriptor: | (3~{R})-1-phenylpyrrolidine-3-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ruza, R.R, Hillier, J, Jones, E.Y. | | Deposit date: | 2020-04-27 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Screening of a Custom-Designed Acid Fragment Library Identifies 1-Phenylpyrroles and 1-Phenylpyrrolidines as Inhibitors of Notum Carboxylesterase Activity.
J.Med.Chem., 63, 2020
|
|
6YUY
 
 | | STRUCTURE OF THE WNT DEACYLASE NOTUM IN COMPLEX WITH A PYRROLE-3-CARBOXYLIC ACID FRAGMENT 471 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-methyl-5-(trifluoromethyl)-1~{H}-pyrrole-3-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Hillier, J, Ruza, R.R, Jones, E.Y. | | Deposit date: | 2020-04-27 | | Release date: | 2020-05-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Screening of a Custom-Designed Acid Fragment Library Identifies 1-Phenylpyrroles and 1-Phenylpyrrolidines as Inhibitors of Notum Carboxylesterase Activity.
J.Med.Chem., 63, 2020
|
|
6YXI
 
 | | Structure of Notum in complex with a 1-(3-Chlorophenyl)-2,5-dimethyl-1H-pyrrole-3-carboxylic acid inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-chlorophenyl)-2,5-dimethyl-pyrrole-3-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Vecchia, L, Jones, E.Y, Ruza, R.R, Hillier, J, Zhao, Y. | | Deposit date: | 2020-05-01 | | Release date: | 2020-08-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Screening of a Custom-Designed Acid Fragment Library Identifies 1-Phenylpyrroles and 1-Phenylpyrrolidines as Inhibitors of Notum Carboxylesterase Activity.
J.Med.Chem., 63, 2020
|
|
6YV4
 
 | | STRUCTURE OF THE WNT DEACYLASE NOTUM IN COMPLEX WITH A PYRROLE-3-CARBOXYLIC ACID FRAGMENT 686 | | Descriptor: | 1,2-ETHANEDIOL, 1-cyclopropyl-2,5-dimethyl-pyrrole-3-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hillier, J, Ruza, R.R, Jones, E.Y. | | Deposit date: | 2020-04-27 | | Release date: | 2020-05-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Screening of a Custom-Designed Acid Fragment Library Identifies 1-Phenylpyrroles and 1-Phenylpyrrolidines as Inhibitors of Notum Carboxylesterase Activity.
J.Med.Chem., 63, 2020
|
|
6YV0
 
 | | STRUCTURE OF THE WNT DEACYLASE NOTUM IN COMPLEX WITH A PYRROLIDINE-3-CARBOXYLIC ACID FRAGMENT 587 | | Descriptor: | (3~{R})-1-(2-chlorophenyl)pyrrolidine-3-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ruza, R.R, Hillier, J, Jones, E.Y. | | Deposit date: | 2020-04-27 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Screening of a Custom-Designed Acid Fragment Library Identifies 1-Phenylpyrroles and 1-Phenylpyrrolidines as Inhibitors of Notum Carboxylesterase Activity.
J.Med.Chem., 63, 2020
|
|
8FIU
 
 | | Potent long-acting inhibitors targeting HIV-1 capsid based on a versatile quinazolin-4-one scaffold | | Descriptor: | 1,2-ETHANEDIOL, HIV-1 capsid, N-[(1S)-1-{(3P)-3-{4-chloro-3-[(methanesulfonyl)amino]-1-methyl-1H-indazol-7-yl}-7-[(2R,6S)-2,6-dimethylmorpholin-4-yl]-4-oxo-3,4-dihydroquinazolin-2-yl}-2-(3,5-difluorophenyl)ethyl]-2-[(3bS,4aR)-3-(difluoromethyl)-5,5-difluoro-3b,4,4a,5-tetrahydro-1H-cyclopropa[3,4]cyclopenta[1,2-c]pyrazol-1-yl]acetamide | | Authors: | Nolte, R.T. | | Deposit date: | 2022-12-16 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Potent Long-Acting Inhibitors Targeting the HIV-1 Capsid Based on a Versatile Quinazolin-4-one Scaffold.
J.Med.Chem., 66, 2023
|
|
9DOD
 
 | | Crystal structure of PRMT5:MEP50 in complex with PRT543 | | Descriptor: | 1,2-ETHANEDIOL, 7-[(3xi,5R)-5-C-(3,4-dichlorophenyl)-3-C-methyl-beta-D-ribofuranosyl]-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Methylosome protein WDR77, ... | | Authors: | Leal, R.A, Pitis, P.M, Dou, Y, Lin, H, Wang, M, Xu, C. | | Deposit date: | 2024-09-18 | | Release date: | 2025-02-12 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Phase Ib study of PRT543, an oral protein arginine methyltransferase 5 (PRMT5) inhibitor, in patients with advanced splicing factor-mutant myeloid malignancies.
Leukemia, 39, 2025
|
|
8SIW
 
 | | Structure of Compound 5 bound to the CHK1 10-point mutant | | Descriptor: | (1S,2S)-N-(7-chloro-6-{1-[(3R,4R)-4-hydroxy-3-methyloxolan-3-yl]piperidin-4-yl}isoquinolin-3-yl)-2-(1-methyl-1H-pyrazol-4-yl)cyclopropane-1-carboxamide, Serine/threonine-protein kinase Chk1 | | Authors: | Palte, R.L. | | Deposit date: | 2023-04-17 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.877 Å) | | Cite: | Discovery of MK-1468: A Potent, Kinome-Selective, Brain-Penetrant Amidoisoquinoline LRRK2 Inhibitor for the Potential Treatment of Parkinson's Disease.
J.Med.Chem., 66, 2023
|
|
8SIV
 
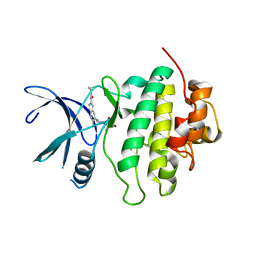 | | Structure of Compound 2 bound to the CHK1 10-point mutant | | Descriptor: | N-(7-chloro-6-{1-[(3R,4R)-4-hydroxy-3-methyloxolan-3-yl]piperidin-4-yl}isoquinolin-3-yl)cyclopropanecarboxamide, Serine/threonine-protein kinase Chk1 | | Authors: | Palte, R.L. | | Deposit date: | 2023-04-17 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.759 Å) | | Cite: | Discovery of MK-1468: A Potent, Kinome-Selective, Brain-Penetrant Amidoisoquinoline LRRK2 Inhibitor for the Potential Treatment of Parkinson's Disease.
J.Med.Chem., 66, 2023
|
|
8SIX
 
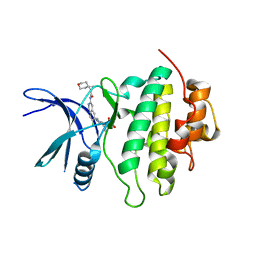 | | Structure of Compound 13 bound to the CHK1 10-point mutant | | Descriptor: | (1S)-N-(7-chloro-6-{4-[(3R,4R)-4-hydroxy-3-methyloxolan-3-yl]piperazin-1-yl}isoquinolin-3-yl)-6-oxaspiro[2.5]octane-1-carboxamide, Serine/threonine-protein kinase Chk1 | | Authors: | Palte, R.L. | | Deposit date: | 2023-04-17 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of MK-1468: A Potent, Kinome-Selective, Brain-Penetrant Amidoisoquinoline LRRK2 Inhibitor for the Potential Treatment of Parkinson's Disease.
J.Med.Chem., 66, 2023
|
|
7U2U
 
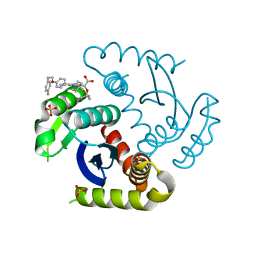 | | CRYSTAL STRUCTURE OF HIV-1 INTEGRASE COMPLEXED WITH Compound-2a AKA (2S)-2-(TERT-BUTOXY)-2-[7-(4,4-DIMETHYLPIPE RIDIN-1-YL)-8-{4-[2-(4-FLUOROPHENYL)ETHOXY]PHENYL}-2,5-DIM ETHYLIMIDAZO[1,2-A]PYRIDIN-6-YL]ACETIC ACID | | Descriptor: | (2S)-tert-butoxy[(4S)-7-(4,4-dimethylpiperidin-1-yl)-8-{4-[2-(4-fluorophenyl)ethoxy]phenyl}-2,5-dimethylimidazo[1,2-a]pyridin-6-yl]acetic acid, Integrase, SULFATE ION | | Authors: | Khan, J.A, lewis, H, Kish, K. | | Deposit date: | 2022-02-24 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.839 Å) | | Cite: | Scaffold modifications to the 4-(4,4-dimethylpiperidinyl) 2,6-dimethylpyridinyl class of HIV-1 allosteric integrase inhibitors.
Bioorg.Med.Chem., 67, 2022
|
|
6NCJ
 
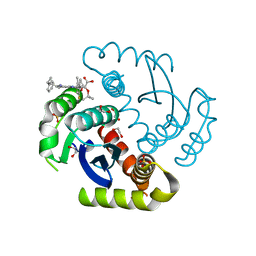 | | Structure of HIV-1 Integrase with potent 5,6,7,8-Tetrahydro-1,6-naphthyridine Derivatives Allosteric Site Inhibitors | | Descriptor: | (2~{S})-2-[4-(8-fluoranyl-5-methyl-3,4-dihydro-2~{H}-chromen-6-yl)-2-methyl-6-[[(1~{S},2~{R})-2-phenylcyclopropyl]methyl]-7,8-dihydro-5~{H}-1,6-naphthyridin-3-yl]-2-[(2-methylpropan-2-yl)oxy]ethanoic acid, 1,2-ETHANEDIOL, Integrase, ... | | Authors: | Nolte, R.T. | | Deposit date: | 2018-12-11 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 5,6,7,8-Tetrahydro-1,6-naphthyridine Derivatives as Potent HIV-1-Integrase-Allosteric-Site Inhibitors.
J. Med. Chem., 62, 2019
|
|
3ZD1
 
 | | STRUCTURE OF THE TWO C-TERMINAL DOMAINS OF COMPLEMENT FACTOR H RELATED PROTEIN 2 | | Descriptor: | 1,2-ETHANEDIOL, COMPLEMENT FACTOR H-RELATED PROTEIN 2 | | Authors: | Caesar, J.J.E, Goicoechea de Jorge, E, Pickering, M.C, Lea, S.M. | | Deposit date: | 2012-11-23 | | Release date: | 2013-03-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dimerization of Complement Factor H-Related Proteins Modulates Complement Activation in Vivo.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7T9H
 
 | | HIV Integrase in complex with Compound-15 | | Descriptor: | (2S)-tert-butoxy[2-methyl-4-(4-methylphenyl)quinolin-3-yl]acetic acid, Integrase, MAGNESIUM ION | | Authors: | Khan, J.A, Lewis, H, Kish, K. | | Deposit date: | 2021-12-19 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Design, Synthesis, and Preclinical Profiling of GSK3739936 (BMS-986180), an Allosteric Inhibitor of HIV-1 Integrase with Broad-Spectrum Activity toward 124/125 Polymorphs.
J.Med.Chem., 65, 2022
|
|
7T9O
 
 | | HIV Integrase in complex with Compound-25 | | Descriptor: | (2S)-tert-butoxy[4-(4,4-dimethylpiperidin-1-yl)-5-{4-[2-(4-fluorophenyl)ethoxy]phenyl}-2,6-dimethylpyridin-3-yl]acetic acid, GLYCEROL, Integrase, ... | | Authors: | Khan, J.A, Lewis, H, Kish, K. | | Deposit date: | 2021-12-19 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design, Synthesis, and Preclinical Profiling of GSK3739936 (BMS-986180), an Allosteric Inhibitor of HIV-1 Integrase with Broad-Spectrum Activity toward 124/125 Polymorphs.
J.Med.Chem., 65, 2022
|
|
3ZD2
 
 | | THE STRUCTURE OF THE TWO N-TERMINAL DOMAINS OF COMPLEMENT FACTOR H RELATED PROTEIN 1 SHOWS FORMATION OF A NOVEL DIMERISATION INTERFACE | | Descriptor: | 1,2-ETHANEDIOL, COMPLEMENT FACTOR H-RELATED PROTEIN 1 | | Authors: | Caesar, J.J.E, Goicoechea de Jorge, E, Pickering, M.C, Lea, S.M. | | Deposit date: | 2012-11-23 | | Release date: | 2013-03-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Dimerization of Complement Factor H-Related Proteins Modulates Complement Activation in Vivo.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6R95
 
 | |
6R96
 
 | |
8FK4
 
 | | Structure of the catalytic domain of Streptococcus mutans GtfB complexed to acarbose in orthorhombic space group P21212 | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, CALCIUM ION, Glucosyltransferase-I, ... | | Authors: | Schormann, N, Deivanayagam, C. | | Deposit date: | 2022-12-20 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | The catalytic domains of Streptococcus mutans glucosyltransferases: a structural analysis.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
8FJ9
 
 | | Structure of the catalytic domain of Streptococcus mutans GtfB in tetragonal space group P4322 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Schormann, N, Deivanayagam, C. | | Deposit date: | 2022-12-19 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The catalytic domains of Streptococcus mutans glucosyltransferases: a structural analysis.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
8FJC
 
 | | Structure of the catalytic domain of Streptococcus mutans GtfB complexed to acarbose in tetragonal space group P4322 | | Descriptor: | 1,2-ETHANEDIOL, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, CALCIUM ION, ... | | Authors: | Schormann, N, Deivanayagam, C. | | Deposit date: | 2022-12-19 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The catalytic domains of Streptococcus mutans glucosyltransferases: a structural analysis.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
8FKL
 
 | |
8FN5
 
 | |
7KIS
 
 | | Crystal structure of Pseudomonas aeruginosa PBP2 in complex with WCK 5153 | | Descriptor: | (2S,5R)-1-formyl-N'-[(3R)-pyrrolidine-3-carbonyl]-5-[(sulfooxy)amino]piperidine-2-carbohydrazide, CHLORIDE ION, Peptidoglycan D,D-transpeptidase MrdA | | Authors: | Rajavel, M, van den Akker, F. | | Deposit date: | 2020-10-24 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.869 Å) | | Cite: | Structural Characterization of Diazabicyclooctane beta-Lactam "Enhancers" in Complex with Penicillin-Binding Proteins PBP2 and PBP3 of Pseudomonas aeruginosa.
Mbio, 12, 2021
|
|
6CPW
 
 | | Discovery of 3(S)-thiomethyl pyrrolidine ERK inhibitors for oncology | | Descriptor: | (3S)-N-[3-(4-fluorophenyl)-1H-indazol-5-yl]-3-(methylsulfanyl)-1-(2-oxo-2-{4-[4-(pyrimidin-2-yl)phenyl]piperazin-1-yl}ethyl)pyrrolidine-3-carboxamide, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Hruza, A, Hruza, A. | | Deposit date: | 2018-03-14 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of 3(S)-thiomethyl pyrrolidine ERK inhibitors for oncology.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
