5J0L
 
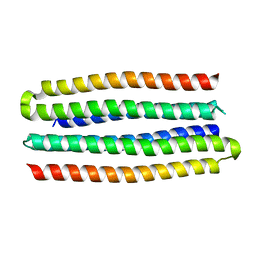 | | De novo design of protein homo-oligomers with modular hydrogen bond network-mediated specificity | | Descriptor: | designed protein 3L6HC2_2 | | Authors: | Sankaran, B, Zwart, P.H, Pereira, J.H, Baker, D, Boyken, S, Chen, Z, Groves, B, Langan, R.A, Oberdorfer, G, Ford, A, Gilmore, J, Xu, C, DiMaio, F, Seelig, G. | | Deposit date: | 2016-03-28 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | De novo design of protein homo-oligomers with modular hydrogen-bond network-mediated specificity.
Science, 352, 2016
|
|
5J0K
 
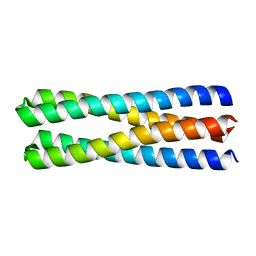 | | De novo design of protein homo-oligomers with modular hydrogen bond network-mediated specificity | | Descriptor: | designed protein 2L4HC2_23 | | Authors: | Sankaran, B, Zwart, P.H, Pereira, J.H, Baker, D, Boyken, S, Chen, Z, Groves, B, Langan, R.A, Oberdorfer, G, Ford, A, Gilmore, J, Xu, C, DiMaio, F, Seelig, G. | | Deposit date: | 2016-03-28 | | Release date: | 2016-05-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | De novo design of protein homo-oligomers with modular hydrogen-bond network-mediated specificity.
Science, 352, 2016
|
|
5J10
 
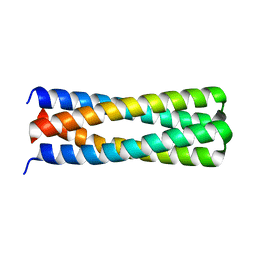 | | De novo design of protein homo-oligomers with modular hydrogen bond network-mediated specificity | | Descriptor: | peptide design 2L4HC2_24 | | Authors: | Sankaran, B, Zwart, P.H, Pereira, J.H, Baker, D, Boyken, S, Chen, Z, Groves, B, Langan, R.A, Oberdorfer, G, Ford, A, Gilmore, J, Xu, C, DiMaio, F, Seelig, G. | | Deposit date: | 2016-03-28 | | Release date: | 2016-05-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | De novo design of protein homo-oligomers with modular hydrogen-bond network-mediated specificity.
Science, 352, 2016
|
|
5J0J
 
 | | De novo design of protein homo-oligomers with modular hydrogen bond network-mediated specificity | | Descriptor: | designed protein 2L6HC3_6 | | Authors: | Sankaran, B, Zwart, P.H, Pereira, J.H, Baker, D, Boyken, S, Chen, Z, Groves, B, Langan, R.A, Oberdorfer, G, Ford, A, Gilmore, J, Xu, C, DiMaio, F, Seelig, G. | | Deposit date: | 2016-03-28 | | Release date: | 2016-05-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.256 Å) | | Cite: | De novo design of protein homo-oligomers with modular hydrogen-bond network-mediated specificity.
Science, 352, 2016
|
|
5J73
 
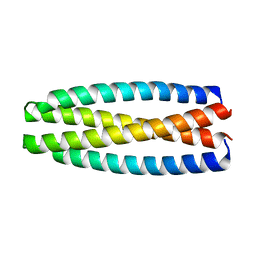 | | De novo design of protein homo-oligomers with modular hydrogen bond network-mediated specificity | | Descriptor: | protein design 2L4HC2_9 | | Authors: | Sankaran, B, Zwart, P.H, Pereira, J.H, Baker, D, Boyken, S, Chen, Z, Groves, B, Langan, R.A, Oberdorfer, G, Ford, A, Gilmore, J, Xu, C, DiMaio, F, Seelig, G. | | Deposit date: | 2016-04-05 | | Release date: | 2016-05-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | De novo design of protein homo-oligomers with modular hydrogen-bond network-mediated specificity.
Science, 352, 2016
|
|
5J2L
 
 | | De novo design of protein homo-oligomers with modular hydrogen bond network-mediated specificity | | Descriptor: | protein design 2L4HC2_11 | | Authors: | Sankaran, B, Zwart, P.H, Pereira, J.H, Baker, D, Boyken, S, Chen, Z, Groves, B, Langan, R.A, Oberdorfer, G, Ford, A, Gilmore, J, Xu, C, DiMaio, F, Seelig, G. | | Deposit date: | 2016-03-29 | | Release date: | 2016-05-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | De novo design of protein homo-oligomers with modular hydrogen-bond network-mediated specificity.
Science, 352, 2016
|
|
5IZS
 
 | | De novo design of protein homo-oligomers with modular hydrogen bond network-mediated specificity | | Descriptor: | Designed protein 5L6HC3_1 | | Authors: | Sankaran, B, Zwart, P.H, Pereira, J.H, Baker, D, Boyken, S, Chen, Z, Oberdorfer, G. | | Deposit date: | 2016-03-25 | | Release date: | 2016-05-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | De novo design of protein homo-oligomers with modular hydrogen-bond network-mediated specificity.
Science, 352, 2016
|
|
8P0Z
 
 | | AP01-S2.3 - a variant of a redesigned transferrin receptor apical domain | | Descriptor: | BORIC ACID, SODIUM ION, Transferrin receptor protein 1, ... | | Authors: | Oberdorfer, G, Grill, B, Bjelic, S, Stoll, D. | | Deposit date: | 2023-05-11 | | Release date: | 2023-09-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Affinity Maturated Transferrin Receptor Apical Domain Blocks Machupo Virus Glycoprotein Binding.
J.Mol.Biol., 435, 2023
|
|
6E9X
 
 | | DHF91 filament | | Descriptor: | DHF91 filament | | Authors: | Lynch, E.M, Shen, H, Fallas, J.A, Kollman, J.M, Baker, D. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | De novo design of self-assembling helical protein filaments.
Science, 362, 2018
|
|
7SKN
 
 | |
7SKP
 
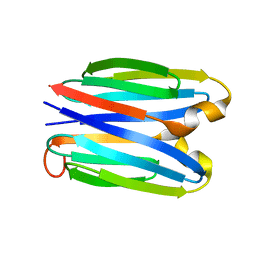 | |
7SKO
 
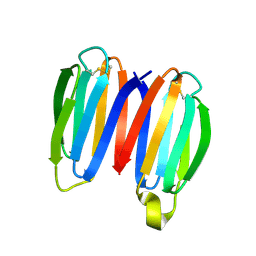 | | De novo synthetic protein DIG8-CC (orthogonal space group) | | Descriptor: | De novo synthetic protein DIG8-CC, MAGNESIUM ION | | Authors: | Mendes, S.R, Eckhard, U, Marcos, E, Gomis-Ruth, F.X. | | Deposit date: | 2021-10-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | De novo design of immunoglobulin-like domains
Nat Commun, 13, 2022
|
|
8AEP
 
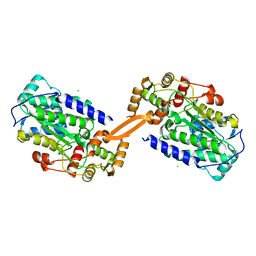 | | Reductase domain of the carboxylate reductase of Neurospora crassa | | Descriptor: | Acetyl-CoA synthetase-like protein, CHLORIDE ION, SULFATE ION | | Authors: | Daniel, B, Schrufer, A, Marlene, L, Sagmeister, T, Pavkov-Keller, T. | | Deposit date: | 2022-07-13 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Reductase Domain of a Fungal Carboxylic Acid Reductase and Its Substrate Scope in Thioester and Aldehyde Reduction.
Acs Catalysis, 12, 2022
|
|
8RO4
 
 | | The crystal structure of 2-hydroxy-3-keto-glucal hydratase AtHYD from A. tumefaciens | | Descriptor: | 2-hydroxy-3-keto-glucal hydratase, MANGANESE (II) ION | | Authors: | Grininger, C, Bitter, J, Pfeiffer, M, Nidetzky, B, Pavkov-Keller, T. | | Deposit date: | 2024-01-11 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Enzyme Machinery for Bacterial Glucoside Metabolism through a Conserved Non-hydrolytic Pathway.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
6NAF
 
 | | De novo designed homo-trimeric amantadine-binding protein | | Descriptor: | (3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-amine, SODIUM ION, amantadine-binding protein | | Authors: | Selvaraj, B, Park, J, Cuneo, M.J, Myles, D.A.A, Baker, D. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (1.923 Å), X-RAY DIFFRACTION | | Cite: | De novo design of a homo-trimeric amantadine-binding protein.
Elife, 8, 2019
|
|
6N9H
 
 | | De novo designed homo-trimeric amantadine-binding protein | | Descriptor: | (3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-amine, SODIUM ION, amantadine-binding protein | | Authors: | Park, J, Baker, D. | | Deposit date: | 2018-12-03 | | Release date: | 2019-12-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.039 Å) | | Cite: | De novo design of a homo-trimeric amantadine-binding protein.
Elife, 8, 2019
|
|
5D79
 
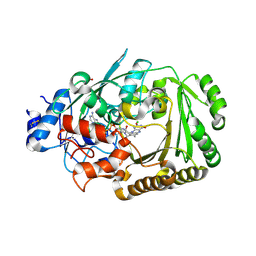 | | Structure of BBE-like #28 from Arabidopsis thaliana | | Descriptor: | Berberine bridge enzyme-like protein, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Daniel, B, Kumar, P, Gruber, K. | | Deposit date: | 2015-08-13 | | Release date: | 2016-06-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Structure of a Berberine Bridge Enzyme-Like Enzyme with an Active Site Specific to the Plant Family Brassicaceae.
Plos One, 11, 2016
|
|
5EIL
 
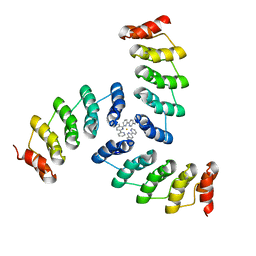 | | Computational design of a high-affinity metalloprotein homotrimer containing a metal chelating non-canonical amino acid | | Descriptor: | FE (III) ION, TRI-05 | | Authors: | Sankaran, B, Zwart, P.H, Mills, J.H, Pereira, J.H, Baker, D. | | Deposit date: | 2015-10-30 | | Release date: | 2016-11-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Computational design of a homotrimeric metalloprotein with a trisbipyridyl core.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5KPH
 
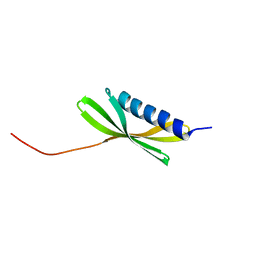 | |
5KPE
 
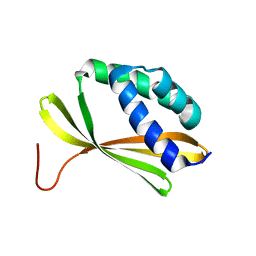 | | Solution NMR Structure of Denovo Beta Sheet Design Protein, Northeast Structural Genomics Consortium (NESG) Target OR664 | | Descriptor: | De novo Beta Sheet Design Protein OR664 | | Authors: | Tang, Y, Liu, G, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2016-07-03 | | Release date: | 2016-09-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
4FQF
 
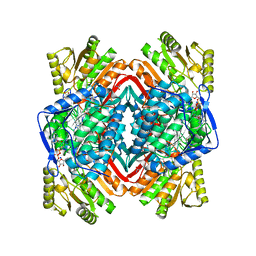 | |
4FR8
 
 | | Crystal structure of human aldehyde dehydrogenase-2 in complex with nitroglycerin | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, Aldehyde dehydrogenase, ... | | Authors: | Lang, B.S, Gruber, K. | | Deposit date: | 2012-06-26 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Vascular Bioactivation of Nitroglycerin by Aldehyde Dehydrogenase-2: REACTION INTERMEDIATES REVEALED BY CRYSTALLOGRAPHY AND MASS SPECTROMETRY.
J.Biol.Chem., 287, 2012
|
|
6E5C
 
 | | Solution NMR structure of a de novo designed double-stranded beta-helix | | Descriptor: | De novo beta protein | | Authors: | Marcos, E, Chidyausiku, T.M, McShan, A, Evangelidis, T, Nerli, S, Sgourakis, N, Tripsianes, K, Baker, D. | | Deposit date: | 2018-07-19 | | Release date: | 2018-11-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | De novo design of a non-local beta-sheet protein with high stability and accuracy.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6E9R
 
 | | DHF46 filament | | Descriptor: | DHF46 filament | | Authors: | Lynch, E.M, Shen, H, Fallas, J.A, Kollman, J.M, Baker, D. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | De novo design of self-assembling helical protein filaments.
Science, 362, 2018
|
|
6E9V
 
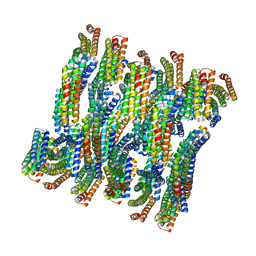 | | DHF79 filament | | Descriptor: | DHF79 filament | | Authors: | Lynch, E.M, Shen, H, Fallas, J.A, Kollman, J.M, Baker, D. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | De novo design of self-assembling helical protein filaments.
Science, 362, 2018
|
|
