4Q5Q
 
 | |
4Q5N
 
 | |
4Q5F
 
 | |
1J57
 
 | | NuiA | | Descriptor: | NuiA | | Authors: | Kirby, T.W, Mueller, G.A, DeRose, E.F, Lebetkin, M.S, Meiss, G, Pingoud, A, London, R.E. | | Deposit date: | 2002-01-17 | | Release date: | 2002-12-04 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The Nuclease A Inhibitor represents a new variation of the rare PR-1 fold.
J.Mol.Biol., 320, 2002
|
|
1KTJ
 
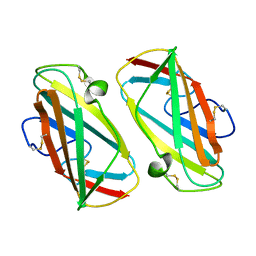 | | X-ray Structure Of Der P 2, The Major House Dust Mite Allergen | | Descriptor: | ALLERGEN DER P 2 | | Authors: | Derewenda, U, Li, J, Derewenda, Z, Dauter, Z, Mueller, G.A, Rule, G.S, Benjamin, D.C. | | Deposit date: | 2002-01-16 | | Release date: | 2002-05-15 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of a major dust mite allergen Der p 2, and its biological implications.
J.Mol.Biol., 318, 2002
|
|
1KTU
 
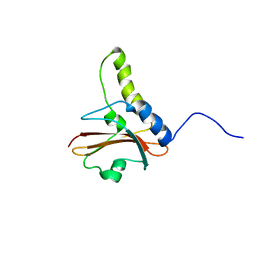 | | NuiA | | Descriptor: | NuiA | | Authors: | Kirby, T.W, Mueller, G.A, DeRose, E.F, Lebetkin, M.S, Meiss, G, Pingoud, A, London, R.E. | | Deposit date: | 2002-01-17 | | Release date: | 2002-12-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The Nuclease A Inhibitor represents a new variation of the rare PR-1 fold.
J.Mol.Biol., 320, 2002
|
|
4EHQ
 
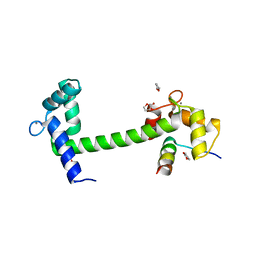 | | Crystal Structure of Calmodulin Binding Domain of Orai1 in Complex with Ca2+/Calmodulin Displays a Unique Binding Mode | | Descriptor: | CALCIUM ION, Calcium release-activated calcium channel protein 1, Calmodulin, ... | | Authors: | Liu, Y, Zheng, X, Mueller, G.A, Sobhany, M, DeRose, E.F, Zhang, Y, London, R.E, Birnbaumer, L. | | Deposit date: | 2012-04-03 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9005 Å) | | Cite: | Crystal structure of calmodulin binding domain of orai1 in complex with ca2+*calmodulin displays a unique binding mode.
J.Biol.Chem., 287, 2012
|
|
4KSE
 
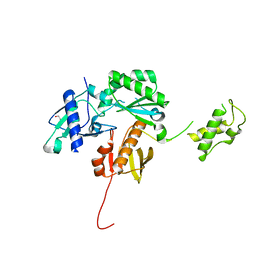 | | Crystal structure of a HIV p51 (219-230) deletion mutant | | Descriptor: | 1,2-ETHANEDIOL, HIV p51 subunit | | Authors: | Zheng, X, Mueller, G.A, Derose, E.F, Pedersen, L.C, Gabel, S.A, Cuneo, M.J, Krahn, J.M, London, R.E. | | Deposit date: | 2013-05-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.677 Å) | | Cite: | Selective unfolding of one Ribonuclease H domain of HIV reverse transcriptase is linked to homodimer formation.
Nucleic Acids Res., 42, 2014
|
|
6XRX
 
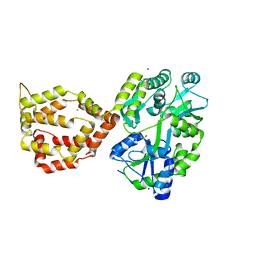 | | Crystal structure of the mosquito protein AZ1 as an MBP fusion | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Maltose/maltodextrin-binding periplasmic protein, ... | | Authors: | Pedersen, L.C, Mueller, G.A, Foo, A.C.Y. | | Deposit date: | 2020-07-14 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The mosquito protein AEG12 displays both cytolytic and antiviral properties via a common lipid transfer mechanism.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2KEW
 
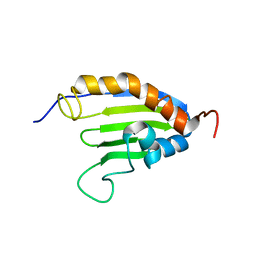 | | The solution structure of Bacillus subtilis SR211 START domain by NMR spectroscopy | | Descriptor: | Uncharacterized protein yndB | | Authors: | Mercier, K.A, Mueller, G.A, Powers, R, Acton, T.B, Ciano, M, Ho, C, Lui, J, Ma, L, Rost, B, Rossi, R, Xiao, R, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-02-05 | | Release date: | 2009-03-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | (1)H, (13)C, and (15)N NMR assignments for the Bacillus subtilis yndB START domain.
Biomol.Nmr Assign., 3, 2009
|
|
2KTE
 
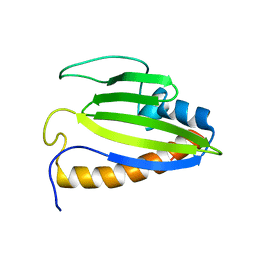 | | The solution structure of Bacillus subtilis, YndB, Northeast Structural Genomics Consoritum Target SR211 | | Descriptor: | Uncharacterized protein yndB | | Authors: | Mercier, K.A, Mueller, G.A, Powers, R, Acton, T.B, Ciano, M, Ho, C, Lui, J, Ma, L, Rost, B, Rossi, R, Montelione, G.T, Xiao, R, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-01-29 | | Release date: | 2010-06-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and function of YndB, an AHSA1 protein from Bacillus subtilis.
Proteins, 78, 2010
|
|
2HTF
 
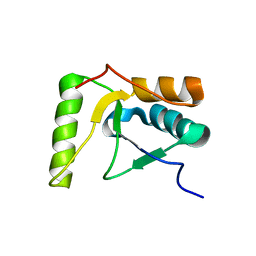 | | The solution structure of the BRCT domain from human polymerase reveals homology with the TdT BRCT domain | | Descriptor: | DNA polymerase mu | | Authors: | DeRose, E.F, Clarkson, M.W, Gilmore, S.A, Ramsden, D.A, Mueller, G.A, London, R.E, Lee, A.L. | | Deposit date: | 2006-07-25 | | Release date: | 2007-02-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of polymerase mu's BRCT Domain reveals an element essential for its role in nonhomologous end joining.
Biochemistry, 46, 2007
|
|
1Z0R
 
 | | Solution Structure of the N-terminal DNA Recognition Domain of the Bacillus subtilis Transcription-State Regulator AbrB | | Descriptor: | Transition state regulatory protein abrB | | Authors: | Bobay, B.G, Andreeva, A, Mueller, G.A, Cavanagh, J, Murzin, A.G. | | Deposit date: | 2005-03-02 | | Release date: | 2005-03-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Revised structure of the AbrB N-terminal domain unifies a diverse superfamily of putative DNA-binding proteins.
Febs Lett., 579, 2005
|
|
1SE7
 
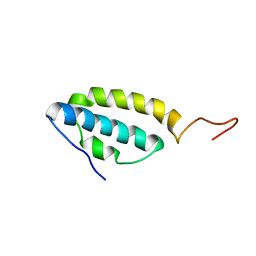 | | Solution structure of the E. coli bacteriophage P1 encoded HOT protein: a homologue of the theta subunit of E. coli DNA polymerase III | | Descriptor: | HOMOLOGUE OF THE THETA SUBUNIT OF DNA POLYMERASE III | | Authors: | DeRose, E.F, Kirby, T.W, Mueller, G.A, Chikova, A.K, Schaaper, R.M, London, R.E. | | Deposit date: | 2004-02-16 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Phage Like It HOT: Solution Structure of the Bacteriophage P1-Encoded HOT Protein, a Homolog of the theta Subunit of E. coli DNA Polymerase III
Structure, 12, 2004
|
|
1O1W
 
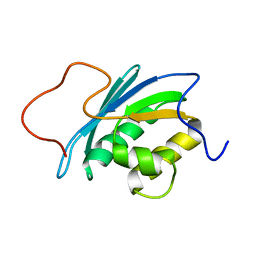 | | SOLUTION STRUCTURE OF THE RNASE H DOMAIN OF THE HIV-1 REVERSE TRANSCRIPTASE IN THE PRESENCE OF MAGNESIUM | | Descriptor: | RIBONUCLEASE H | | Authors: | Pari, K, Mueller, G.A, Derose, E.F, Kirby, T.W, London, R.E. | | Deposit date: | 2003-02-12 | | Release date: | 2003-02-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Rnase H Domain of the HIV-1 Reverse Transcriptase in the Presence of Magnesium
Biochemistry, 42, 2003
|
|
1NZP
 
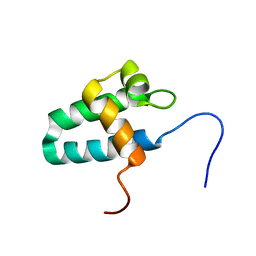 | | Solution Structure of the Lyase Domain of Human DNA Polymerase Lambda | | Descriptor: | DNA polymerase lambda | | Authors: | DeRose, E.F, Kirby, T.W, Mueller, G.A, Bebenek, K, Garcia-Diaz, M, Blanco, L, Kunkel, T.A, London, R.E. | | Deposit date: | 2003-02-19 | | Release date: | 2003-08-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Lyase Domain of Human DNA Polymerase Lambda
Biochemistry, 42, 2003
|
|
1PV3
 
 | | NMR Solution Structure of the Avian FAT-domain of Focal Adhesion Kinase | | Descriptor: | Focal adhesion kinase 1 | | Authors: | Prutzman, K.C, Gao, G, King, M.L, Iyer, V.V, Mueller, G.A, Schaller, M.D, Campbell, S.L. | | Deposit date: | 2003-06-26 | | Release date: | 2004-05-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The Focal Adhesion Targeting Domain of Focal Adhesion Kinase Contains a Hinge Region that Modulates Tyrosine 926 Phosphorylation.
STRUCTURE, 12, 2004
|
|
7LXK
 
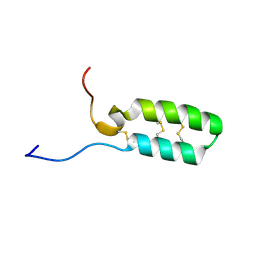 | |
7U65
 
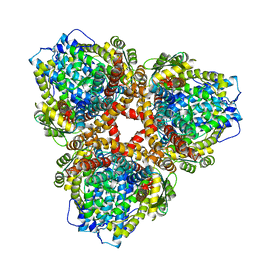 | | Structure of E. coli dGTPase bound to T7 bacteriophage protein Gp1.2 | | Descriptor: | Deoxyguanosinetriphosphate triphosphohydrolase, Inhibitor of dGTPase | | Authors: | Klemm, B.P, Hsu, A.L, Borgnia, M.J, Schaaper, R.M. | | Deposit date: | 2022-03-03 | | Release date: | 2022-08-31 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism by which T7 bacteriophage protein Gp1.2 inhibits Escherichia coli dGTPase.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7U67
 
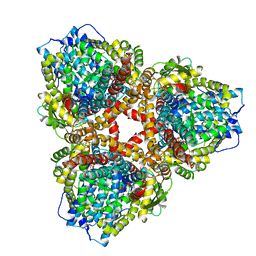 | | Structure of E. coli dGTPase bound to T7 bacteriophage protein Gp1.2 and GTP | | Descriptor: | Deoxyguanosinetriphosphate triphosphohydrolase, GUANOSINE-5'-TRIPHOSPHATE, Inhibitor of dGTPase, ... | | Authors: | Klemm, B.P, Hsu, A.L, Borgnia, M.J, Schaaper, R.M. | | Deposit date: | 2022-03-03 | | Release date: | 2022-08-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Mechanism by which T7 bacteriophage protein Gp1.2 inhibits Escherichia coli dGTPase.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6OY4
 
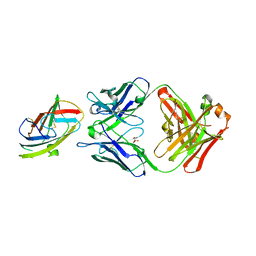 | | Crystal structure of complex between recombinant Der p 2.0103 and Fab fragment of 7A1 | | Descriptor: | Der p 2 variant 3, Fab fragment of IgG, HEAVY CHAIN, ... | | Authors: | Kapingidza, A.B, Offermann, L.R, Glesner, J, Wunschmann, S, Vailes, L.D, Chapman, M.D.C, Pomes, A, Chruszcz, M. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A Human IgE Antibody Binding Site on Der p 2 for the Design of a Recombinant Allergen for Immunotherapy.
J Immunol., 203, 2019
|
|
7U66
 
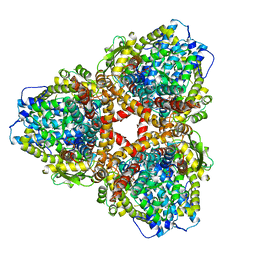 | | Structure of E. coli dGTPase bound to T7 bacteriophage protein Gp1.2 and dGTP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Deoxyguanosinetriphosphate triphosphohydrolase, Inhibitor of dGTPase, ... | | Authors: | Klemm, B.P, Dillard, L.B, Borgnia, M.J, Schaaper, R.M. | | Deposit date: | 2022-03-03 | | Release date: | 2022-08-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism by which T7 bacteriophage protein Gp1.2 inhibits Escherichia coli dGTPase.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
2MC9
 
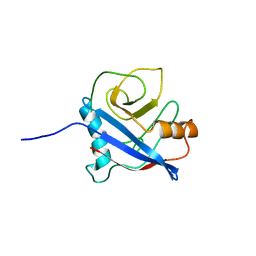 | | Cat r 1 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase | | Authors: | Mueller, G, London, R, Ghosh, D. | | Deposit date: | 2013-08-16 | | Release date: | 2014-06-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Primary identification, biochemical characterization, and immunologic properties of the allergenic pollen cyclophilin cat R 1.
J.Biol.Chem., 289, 2014
|
|
2LE0
 
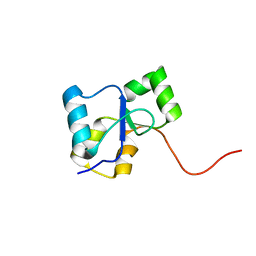 | | PARP BRCT Domain | | Descriptor: | Poly [ADP-ribose] polymerase 1 | | Authors: | Mueller, G, Loeffler, P, Cuneo, M, Derose, E, London, R. | | Deposit date: | 2011-06-03 | | Release date: | 2011-10-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural studies of the PARP-1 BRCT domain.
Bmc Struct.Biol., 11, 2011
|
|
5TVP
 
 | |
