7SFL
 
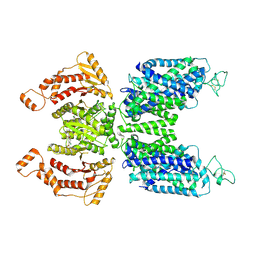 | | Human NKCC1 state Fu-II | | Descriptor: | 5-(AMINOSULFONYL)-4-CHLORO-2-[(2-FURYLMETHYL)AMINO]BENZOIC ACID, Solute carrier family 12 member 2 | | Authors: | Moseng, M.A. | | Deposit date: | 2021-10-04 | | Release date: | 2022-09-28 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Inhibition mechanism of NKCC1 involves the carboxyl terminus and long-range conformational coupling.
Sci Adv, 8, 2022
|
|
7SMP
 
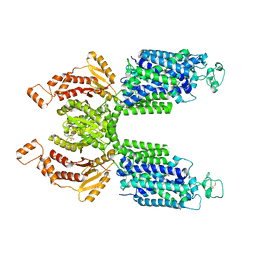 | | CryoEM structure of NKCC1 Bu-I | | Descriptor: | 3-(butylamino)-4-phenoxy-5-sulfamoylbenzoic acid, POTASSIUM ION, Solute carrier family 12 member 2 | | Authors: | Moseng, M.A. | | Deposit date: | 2021-10-26 | | Release date: | 2022-09-28 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Inhibition mechanism of NKCC1 involves the carboxyl terminus and long-range conformational coupling.
Sci Adv, 8, 2022
|
|
8STE
 
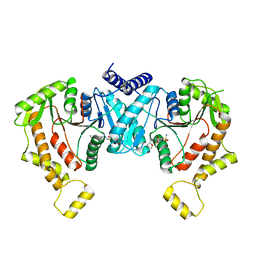 | | Cryo-EM structure of NKCC1 Fu_CTD | | Descriptor: | 5-(AMINOSULFONYL)-4-CHLORO-2-[(2-FURYLMETHYL)AMINO]BENZOIC ACID, Solute carrier family 12 member 2 | | Authors: | Moseng, M.A. | | Deposit date: | 2023-05-10 | | Release date: | 2023-05-17 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Inhibition mechanism of NKCC1 involves the carboxyl terminus and long-range conformational coupling.
Sci Adv, 8, 2022
|
|
7N6B
 
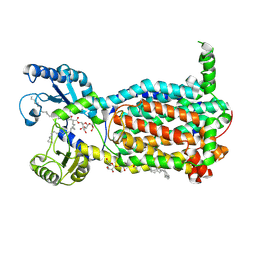 | | Structure of MmpL3 reconstituted into lipid nanodisc in the TMM bound state | | Descriptor: | 6-O-[(2S)-2-{(1S)-18-[(1R,2R)-2-hexylcyclopropyl]-1-hydroxyoctadecyl}tricosanoyl]-alpha-D-glucopyranosyl alpha-D-glucopyranoside, MmpL3 transporter | | Authors: | Su, C.C, Yu, E. | | Deposit date: | 2021-06-08 | | Release date: | 2021-09-01 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structures of the mycobacterial membrane protein MmpL3 reveal its mechanism of lipid transport.
Plos Biol., 19, 2021
|
|
7UZM
 
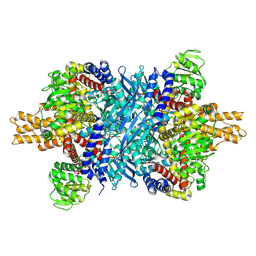 | | Glutamate dehydrogenase 1 from human liver | | Descriptor: | Glutamate dehydrogenase 1, mitochondrial | | Authors: | Zhang, Z. | | Deposit date: | 2022-05-09 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | High-resolution structural-omics of human liver enzymes.
Cell Rep, 42, 2023
|
|
7N3N
 
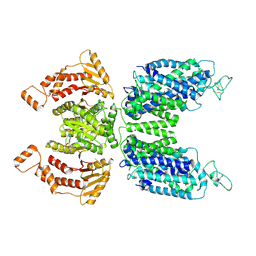 | | CryoEM structure of human NKCC1 state Fu-I | | Descriptor: | 5-(AMINOSULFONYL)-4-CHLORO-2-[(2-FURYLMETHYL)AMINO]BENZOIC ACID, Solute carrier family 12 member 2 | | Authors: | Moseng, M.A. | | Deposit date: | 2021-06-01 | | Release date: | 2022-09-28 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Inhibition mechanism of NKCC1 involves the carboxyl terminus and long-range conformational coupling.
Sci Adv, 8, 2022
|
|
7K8C
 
 | |
7K8D
 
 | |
7K8A
 
 | |
7K8B
 
 | |
7K7M
 
 | | Crystal Structure of a membrane protein | | Descriptor: | Drug exporters of the RND superfamily-like protein, alpha-D-glucopyranose-(1-1)-6-O-decanoyl-alpha-D-glucopyranose | | Authors: | Su, C.-C. | | Deposit date: | 2020-09-23 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Structures of the mycobacterial membrane protein MmpL3 reveal its mechanism of lipid transport.
Plos Biol., 19, 2021
|
|
7MXO
 
 | | CryoEM structure of human NKCC1 | | Descriptor: | CHLORIDE ION, POTASSIUM ION, Solute carrier family 12 member 2 | | Authors: | Moseng, M.A. | | Deposit date: | 2021-05-19 | | Release date: | 2022-09-28 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Inhibition mechanism of NKCC1 involves the carboxyl terminus and long-range conformational coupling.
Sci Adv, 8, 2022
|
|
8EKY
 
 | | Cryo-EM structure of the human PRDX4-ErP46 complex | | Descriptor: | Peroxiredoxin-4, Thioredoxin domain-containing protein 5 | | Authors: | Su, C.C. | | Deposit date: | 2022-09-22 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | High-resolution structural-omics of human liver enzymes.
Cell Rep, 42, 2023
|
|
8EOJ
 
 | | Microsomal triglyceride transfer protein | | Descriptor: | Microsomal triglyceride transfer protein large subunit, Protein disulfide-isomerase | | Authors: | Zhang, Z. | | Deposit date: | 2022-10-03 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | High-resolution structural-omics of human liver enzymes.
Cell Rep, 42, 2023
|
|
8ENE
 
 | |
8EKW
 
 | | Cryo-EM structure of human PRDX4 | | Descriptor: | Peroxiredoxin-4 | | Authors: | Su, C.C. | | Deposit date: | 2022-09-22 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | High-resolution structural-omics of human liver enzymes.
Cell Rep, 42, 2023
|
|
8EOR
 
 | | Liver carboxylesterase 1 | | Descriptor: | ETHYL ACETATE, Liver carboxylesterase 1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Zhang, Z, Yu, E. | | Deposit date: | 2022-10-04 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | High-resolution structural-omics of human liver enzymes.
Cell Rep, 42, 2023
|
|
8EM2
 
 | |
6WTI
 
 | | The Cryo-EM structure of the ubiquinol oxidase from Escherichia coli | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome o ubiquinol oxidase, ... | | Authors: | Su, C.-C. | | Deposit date: | 2020-05-02 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | A 'Build and Retrieve' methodology to simultaneously solve cryo-EM structures of membrane proteins.
Nat.Methods, 18, 2021
|
|
7KF6
 
 | |
7KF8
 
 | |
7KF5
 
 | |
7KF7
 
 | |
7JZ6
 
 | |
7JZH
 
 | |
