5CTV
 
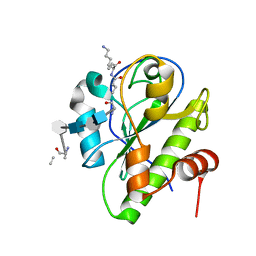 | | Catalytic domain of LytA, the major autolysin of Streptococcus pneumoniae, (C60A, H133A, C136A mutant) complexed with peptidoglycan fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-3-O-[(1R)-1-carboxyethyl]-2-deoxy-beta-D-glucopyranoside-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-3-O-[(1R)-1-carboxyethyl]-2-deoxy-beta-D-glucopyranoside, Autolysin, fragment of peptidoglycan | | Authors: | Achour, A, Sandalova, T, Mellroth, P. | | Deposit date: | 2015-07-24 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | The crystal structure of the major pneumococcal autolysin LytA in complex with a large peptidoglycan fragment reveals the pivotal role of glycans for lytic activity.
Mol.Microbiol., 101, 2016
|
|
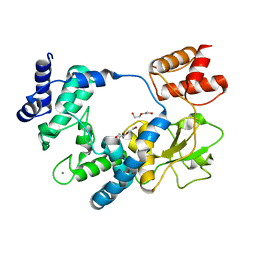
5ANZ
 
 | |
4IWT
 
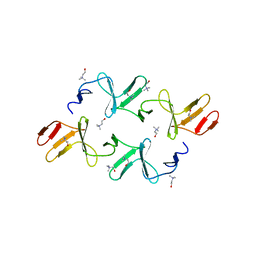 | |
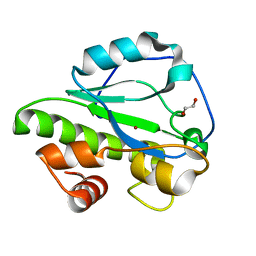
4IVV
 
 | |
5OAU
 
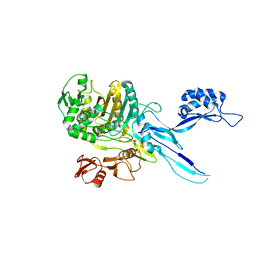 | |
5OJ1
 
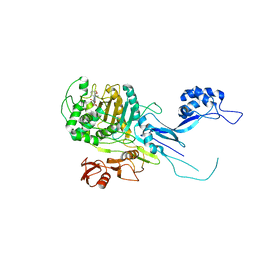 | | Penicillin Binding Protein 2x (PBP2x) from S.pneumoniae in complex with Oxacillin and a tetrasaccharide | | Descriptor: | (2R,4S)-5,5-dimethyl-2-[(1R)-1-{[(5-methyl-3-phenyl-1,2-oxazol-4-yl)carbonyl]amino}-2-oxoethyl]-1,3-thiazolidine-4-carb oxylic acid, Penicillin-binding protein 2X, SODIUM ION | | Authors: | Bernardo-Garcia, N, Hermoso, J.A. | | Deposit date: | 2017-07-20 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Allostery, Recognition of Nascent Peptidoglycan, and Cross-linking of the Cell Wall by the Essential Penicillin-Binding Protein 2x of Streptococcus pneumoniae.
ACS Chem. Biol., 13, 2018
|
|
5OIZ
 
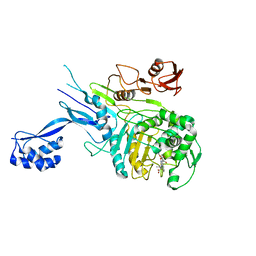 | | Penicillin-Binding Protein 2X (PBP2X) from Streptococcus pneumoniae in complex with oxacillin | | Descriptor: | (2R,4S)-5,5-dimethyl-2-[(1R)-1-{[(5-methyl-3-phenyl-1,2-oxazol-4-yl)carbonyl]amino}-2-oxoethyl]-1,3-thiazolidine-4-carb oxylic acid, Penicillin-binding protein 2X | | Authors: | Bernardo-Garcia, N, Hermoso, J.A. | | Deposit date: | 2017-07-20 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Allostery, Recognition of Nascent Peptidoglycan, and Cross-linking of the Cell Wall by the Essential Penicillin-Binding Protein 2x of Streptococcus pneumoniae.
ACS Chem. Biol., 13, 2018
|
|
7LAM
 
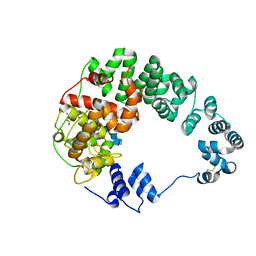 | | Crystal structure of Campylobacter jejuni Cj0843c lytic transglycosylase in complex with N,N',N''-triacetylchitotriose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CITRIC ACID, ... | | Authors: | van den Akker, F, Kumar, V. | | Deposit date: | 2021-01-06 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Turnover Chemistry and Structural Characterization of the Cj0843c Lytic Transglycosylase of Campylobacter jejuni .
Biochemistry, 60, 2021
|
|
7LAQ
 
 | |
7O4C
 
 | |
7O49
 
 | |
7O4A
 
 | |
7O4B
 
 | |
2J8G
 
 | |
2J8F
 
 | |
2IXU
 
 | |
2IXV
 
 | |
4K91
 
 | | Crystal structure of Penicillin-Binding Protein 5 (PBP5) from Pseudomonas aeruginosa in apo state | | Descriptor: | D-ala-D-ala-carboxypeptidase, SUCCINIC ACID | | Authors: | Smith, J, Toth, M, Vakulenko, S, Mobashery, S, Chen, Y. | | Deposit date: | 2013-04-19 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural analysis of the role of Pseudomonas aeruginosa penicillin-binding protein 5 in beta-lactam resistance.
Antimicrob.Agents Chemother., 57, 2013
|
|
3Q82
 
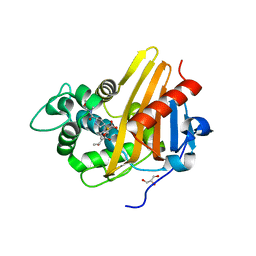 | | Meropenem acylated BlaR1 sensor domain from Staphylococcus aureus | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase regulatory protein BlaR1, GLYCEROL | | Authors: | Borbulevych, O.Y, Mobashery, S, Baker, B.M. | | Deposit date: | 2011-01-05 | | Release date: | 2011-07-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Lysine Nzeta-decarboxylation switch and activation of the beta-lactam sensor domain of BlaR1 protein of methicillin-resistant Staphylococcus aureus.
J.Biol.Chem., 286, 2011
|
|
3Q81
 
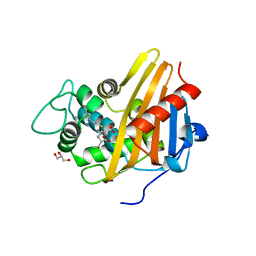 | | Imipenem acylated BlaR1 sensor domain from Staphylococcus aureus | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase regulatory protein BlaR1, GLYCEROL | | Authors: | Borbulevych, O.Y, Mobashery, S, Baker, B.M. | | Deposit date: | 2011-01-05 | | Release date: | 2011-07-20 | | Last modified: | 2013-01-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Lysine Nzeta-decarboxylation switch and activation of the beta-lactam sensor domain of BlaR1 protein of methicillin-resistant Staphylococcus aureus.
J.Biol.Chem., 286, 2011
|
|
3Q7Z
 
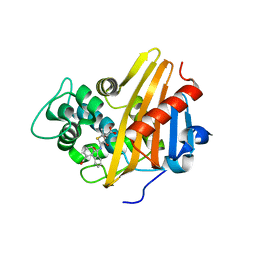 | | CBAP-acylated BlaR1 sensor domain from Staphylococcus aureus | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2'-carboxybiphenyl-2-yl)carbonyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Beta-lactamase regulatory protein BlaR1 | | Authors: | Borbulevych, O.Y, Mobashery, S, Baker, B.M. | | Deposit date: | 2011-01-05 | | Release date: | 2011-07-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Lysine Nzeta-decarboxylation switch and activation of the beta-lactam sensor domain of BlaR1 protein of methicillin-resistant Staphylococcus aureus.
J.Biol.Chem., 286, 2011
|
|
3Q7V
 
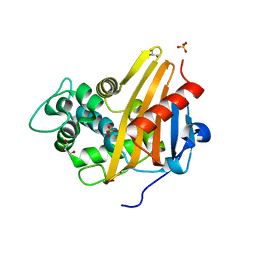 | |
5LY7
 
 | | Crystal structure of NagZ H174A mutant from Pseudomonas aeruginosa in complex with the inhibitor 2-acetamido-1,2-dideoxynojirimycin | | Descriptor: | 2-ACETAMIDO-1,2-DIDEOXYNOJIRMYCIN, Beta-hexosaminidase, DI(HYDROXYETHYL)ETHER | | Authors: | Acebron, I, Artola-Recolons, C, Mahasenan, K, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2016-09-25 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Catalytic Cycle of the N-Acetylglucosaminidase NagZ from Pseudomonas aeruginosa.
J. Am. Chem. Soc., 139, 2017
|
|
5G2M
 
 | | Crystal structure of NagZ from Pseudomonas aeruginosa in complex with N-acetylglucosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-HEXOSAMINIDASE | | Authors: | Acebron, I, Artola-Recolons, C, Mahasenan, K, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2016-04-09 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Catalytic Cycle of the N-Acetylglucosaminidase NagZ from Pseudomonas aeruginosa.
J. Am. Chem. Soc., 139, 2017
|
|
5G5K
 
 | | Crystal structure of NagZ from Pseudomonas aeruginosa in complex with the inhibitor 2-acetamido-1,2-dideoxynojirimycin | | Descriptor: | 2-ACETAMIDO-1,2-DIDEOXYNOJIRMYCIN, BETA-HEXOSAMINIDASE | | Authors: | Acebron, I, Artola-Recolons, C, Mahasenan, K, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2016-05-25 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Catalytic Cycle of the N-Acetylglucosaminidase NagZ from Pseudomonas aeruginosa.
J. Am. Chem. Soc., 139, 2017
|
|
