5O5G
 
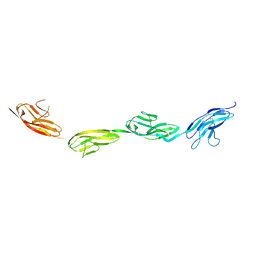 | | Robo1 Ig1 to 4 crystal form 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Roundabout homolog 1 | | Authors: | Aleksandrova, N, Gutsche, I, Kandiah, E, Avilov, S.V, Petoukhov, M.V, Seiradake, E, McCarthy, A.A. | | Deposit date: | 2017-06-01 | | Release date: | 2018-01-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Robo1 Forms a Compact Dimer-of-Dimers Assembly.
Structure, 26, 2018
|
|
5OPE
 
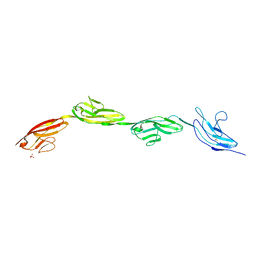 | | Robo1 Ig1-4 crystals form 2 | | Descriptor: | PHOSPHATE ION, Roundabout homolog 1 | | Authors: | Aleksandrova, N, Gutsche, I, Kandiah, E, Avilov, S.V, Petoukhov, M.V, Seiradake, E, McCarthy, A.A. | | Deposit date: | 2017-08-09 | | Release date: | 2018-01-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Robo1 Forms a Compact Dimer-of-Dimers Assembly.
Structure, 26, 2018
|
|
6GGS
 
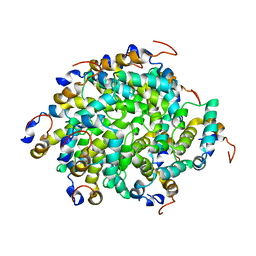 | | Structure of RIP2 CARD filament | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 2 | | Authors: | Pellegrini, E, Cusack, S, Desfosses, A, Schoehn, G, Malet, H, Gutsche, I, Sachse, C, Hons, M. | | Deposit date: | 2018-05-03 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | RIP2 filament formation is required for NOD2 dependent NF-kappa B signalling.
Nat Commun, 9, 2018
|
|
6GK2
 
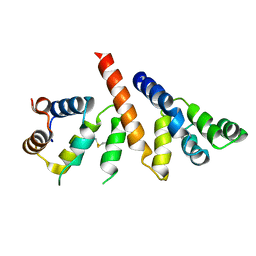 | | Helical reconstruction of BCL10 CARD and MALT1 DEATH DOMAIN complex | | Descriptor: | B-cell lymphoma/leukemia 10, Mucosa-associated lymphoid tissue lymphoma translocation protein 1 | | Authors: | Schlauderer, F, Desfosses, A, Gutsche, I, Hopfner, K.P, Lammens, K. | | Deposit date: | 2018-05-18 | | Release date: | 2018-10-31 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Molecular architecture and regulation of BCL10-MALT1 filaments.
Nat Commun, 9, 2018
|
|
6H5S
 
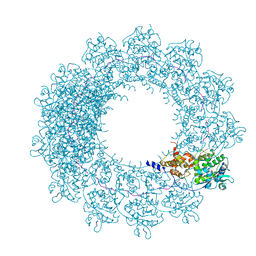 | | Cryo-EM map of in vitro assembled Measles virus N into nucleocapsid-like particles (NCLPs) bound to viral genomic 5-prime RNA hexamers. | | Descriptor: | Nucleocapsid, RNA (5'-R(*AP*CP*CP*AP*GP*A)-3') | | Authors: | Desfosses, A, Milles, S, Ringkjobing Jensen, M, Guseva, S, Colletier, J.P, Maurin, D, Schoehn, G, Gutsche, I, Ruigrok, R, Blackledge, M. | | Deposit date: | 2018-07-25 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Assembly and cryo-EM structures of RNA-specific measles virus nucleocapsids provide mechanistic insight into paramyxoviral replication.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6H5Q
 
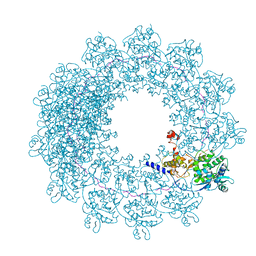 | | Cryo-EM structure of in vitro assembled Measles virus N into nucleocapsid-like particles (NCLPs) bound to polyA RNA hexamers. | | Descriptor: | Nucleocapsid, RNA (5'-R(*AP*AP*AP*AP*AP*A)-3') | | Authors: | Desfosses, A, Milles, S, Ringkjobing Jensen, M, Guseva, S, Colletier, J, Maurin, D, Schoehn, G, Gutsche, I, Ruigrok, R, Blackledge, M. | | Deposit date: | 2018-07-25 | | Release date: | 2019-03-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Assembly and cryo-EM structures of RNA-specific measles virus nucleocapsids provide mechanistic insight into paramyxoviral replication.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5O5I
 
 | | Robo1 Ig5 | | Descriptor: | Roundabout homolog 1 | | Authors: | Aleksandrova, N, Gutsche, I, Kandiah, E, Avilov, S.V, Petoukhov, M.V, Seiradake, E, McCarthy, A.A. | | Deposit date: | 2017-06-01 | | Release date: | 2018-01-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Robo1 Forms a Compact Dimer-of-Dimers Assembly.
Structure, 26, 2018
|
|
7PK6
 
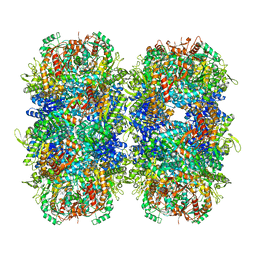 | | Providencia stuartii Arginine decarboxylase (Adc), stack structure | | Descriptor: | Biodegradative arginine decarboxylase | | Authors: | Jessop, M, Desfosses, A, Bacia-Verloop, M, Gutsche, I. | | Deposit date: | 2021-08-25 | | Release date: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (2.15 Å) | | Cite: | Structural and biochemical characterisation of the Providencia stuartii arginine decarboxylase shows distinct polymerisation and regulation.
Commun Biol, 5, 2022
|
|
7P9B
 
 | | Providencia stuartii Arginine decarboxylase (Adc), decamer structure | | Descriptor: | Biodegradative arginine decarboxylase | | Authors: | Jessop, M, Desfosses, A, Bacia-Verloop, M, Gutsche, I. | | Deposit date: | 2021-07-26 | | Release date: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Structural and biochemical characterisation of the Providencia stuartii arginine decarboxylase shows distinct polymerisation and regulation.
Commun Biol, 5, 2022
|
|
7PQH
 
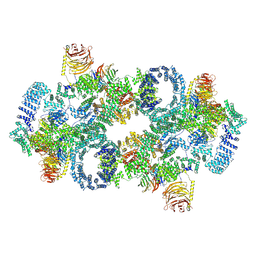 | | Cryo-EM structure of Saccharomyces cerevisiae TOROID (TORC1 Organized in Inhibited Domains). | | Descriptor: | Serine/threonine-protein kinase TOR2, Target of rapamycin complex 1 subunit KOG1,Target of rapamycin complex 1 subunit Kog1, Target of rapamycin complex subunit LST8 | | Authors: | Felix, J, Prouteau, M, Bourgoint, C, Bonadei, L, Desfosses, A, Gabus, C, Sadian, Y, Savvides, S.N, Gutsche, I, Loewith, R. | | Deposit date: | 2021-09-17 | | Release date: | 2023-01-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | EGOC inhibits TOROID polymerization by structurally activating TORC1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6I2N
 
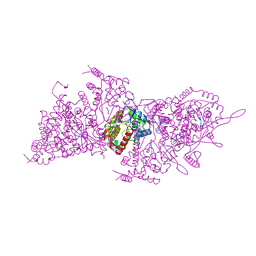 | | Helical RNA-bound Hantaan virus nucleocapsid | | Descriptor: | Nucleoprotein, RNA (5'-R(P*UP*UP*U)-3') | | Authors: | Arragain, B, Reguera, J, Desfosses, A, Gutsche, I, Schoehn, G, Malet, H. | | Deposit date: | 2018-11-01 | | Release date: | 2019-01-23 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | High resolution cryo-EM structure of the helical RNA-bound Hantaan virus nucleocapsid reveals its assembly mechanisms.
Elife, 8, 2019
|
|
6SZA
 
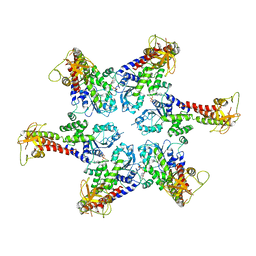 | |
6SZB
 
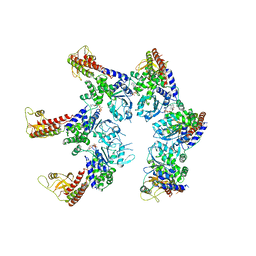 | |
3QS7
 
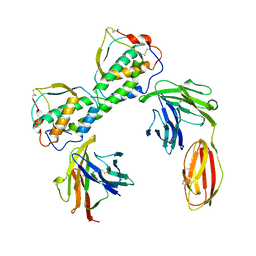 | |
3QS9
 
 | |
5D22
 
 | |
9GNS
 
 | | X-ray structure of Human holo aromatic L-amino acid decarboxylase (AADC) complex with Carbidopa at physiological pH | | Descriptor: | Aromatic-L-amino-acid decarboxylase, CARBIDOPA, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Perduca, M, Bisello, G, Bertoldi, M. | | Deposit date: | 2024-09-04 | | Release date: | 2025-05-14 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | alpha-Hydrazino Acids Inhibit Pyridoxal Phosphate-Dependent Decarboxylases via "Catalytically Correct" Ketoenamine Tautomers: A Special Motif for Chemical Biology and Drug Discovery?
Acs Catalysis, 15, 2025
|
|
6CVZ
 
 | | Crystal structure of the WD40-repeat of RFWD3 | | Descriptor: | E3 ubiquitin-protein ligase RFWD3, MAGNESIUM ION | | Authors: | DONG, A, LOPPNAU, P, SEITOVA, A, HUTCHINSON, A, TEMPEL, W, WEI, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, TONG, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-29 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Target highlights in CASP13: Experimental target structures through the eyes of their authors.
Proteins, 87, 2019
|
|
7CCH
 
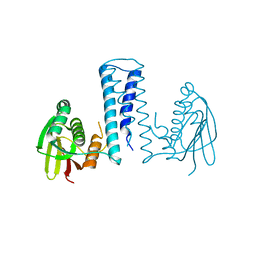 | | Acinetobacter baumannii histidine kinase AdeS | | Descriptor: | AdeS | | Authors: | Wen, Y, Felix, J. | | Deposit date: | 2020-06-17 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.848 Å) | | Cite: | Proteolysis and multimerization regulate signaling along the two-component regulatory system AdeRS.
Iscience, 24, 2021
|
|
7CCI
 
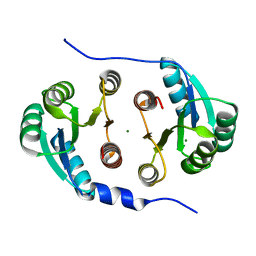 | |
3N75
 
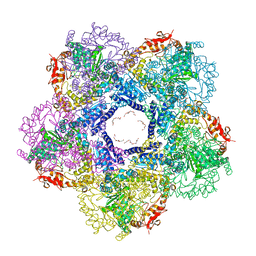 | | X-ray Crystal Structure of the Escherichia coli Inducible Lysine Decarboxylase LdcI | | Descriptor: | GLYCEROL, GUANOSINE-5',3'-TETRAPHOSPHATE, HEXAETHYLENE GLYCOL, ... | | Authors: | Kanjee, U, Alexopoulos, E, Pai, E.F, Houry, W.A. | | Deposit date: | 2010-05-26 | | Release date: | 2011-02-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Linkage between the bacterial acid stress and stringent responses: the structure of the inducible lysine decarboxylase.
Embo J., 30, 2011
|
|
3NBX
 
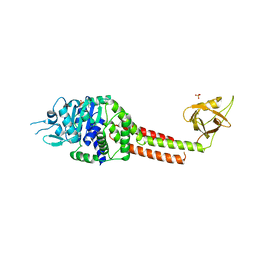 | |
5D28
 
 | |
3UEZ
 
 | |
3UF5
 
 | | Crystal structure of the mouse Colony-Stimulating Factor 1 (mCSF-1) cytokine | | Descriptor: | CALCIUM ION, Macrophage colony-stimulating factor 1 | | Authors: | Elegheert, J, Bracke, N, Bekaert, A, Savvides, S.N. | | Deposit date: | 2011-10-31 | | Release date: | 2012-08-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Allosteric competitive inactivation of hematopoietic CSF-1 signaling by the viral decoy receptor BARF1
Nat.Struct.Mol.Biol., 19, 2012
|
|
