1U5Q
 
 | | Crystal Structure of the TAO2 Kinase Domain: Activation and Specifity of a Ste20p MAP3K | | Descriptor: | CALCIUM ION, serine/threonine protein kinase TAO2 | | Authors: | Zhou, T, Raman, M, Gao, Y, Earnest, S, Chen, Z, Machius, M, Cobb, M.H, Goldsmith, E.J. | | Deposit date: | 2004-07-28 | | Release date: | 2004-10-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the TAO2 Kinase Domain; Activation and Specificity of a Ste20p MAP3K.
STRUCTURE, 12, 2004
|
|
1SEK
 
 | | THE STRUCTURE OF ACTIVE SERPIN K FROM MANDUCA SEXTA AND A MODEL FOR SERPIN-PROTEASE COMPLEX FORMATION | | Descriptor: | SERPIN K | | Authors: | Li, J, Wang, Z, Canagarajah, B, Jiang, H, Kanost, M, Goldsmith, E.J. | | Deposit date: | 1998-03-06 | | Release date: | 1999-03-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of active serpin 1K from Manduca sexta.
Structure Fold.Des., 7, 1999
|
|
1U5R
 
 | | Crystal Structure of the TAO2 Kinase Domain: Activation and Specifity of a Ste20p MAP3K | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Zhou, T, Raman, M, Gao, Y, Earnest, S, Chen, Z, Machius, M, Cobb, M.H, Goldsmith, E.J. | | Deposit date: | 2004-07-28 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the TAO2 Kinase Domain; Activation and Specificity of a Ste20p MAP3K.
Structure, 12, 2004
|
|
1SZR
 
 | | A Dimer interface mutant of ornithine decarboxylase reveals structure of gem diamine intermediate | | Descriptor: | N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], N~2~-({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)-D-ORNITHINE, Ornithine decarboxylase, ... | | Authors: | Jackson, L.K, Baldwin, J, Goldsmith, E.J, Phillips, M.A. | | Deposit date: | 2004-04-06 | | Release date: | 2004-10-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Multiple active site conformations revealed by distant site mutation in ornithine decarboxylase
Biochemistry, 43, 2004
|
|
1K9O
 
 | | CRYSTAL STRUCTURE OF MICHAELIS SERPIN-TRYPSIN COMPLEX | | Descriptor: | ALASERPIN, TRYPSIN II ANIONIC | | Authors: | Ye, S, Cech, A.L, Belmares, R, Bergstrom, R.C, Tong, Y, Corey, D.R, Kanost, M.R, Goldsmith, E.J. | | Deposit date: | 2001-10-29 | | Release date: | 2001-11-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of a Michaelis serpin-protease complex.
Nat.Struct.Biol., 8, 2001
|
|
1LEW
 
 | | CRYSTAL STRUCTURE OF MAP KINASE P38 COMPLEXED TO THE DOCKING SITE ON ITS NUCLEAR SUBSTRATE MEF2A | | Descriptor: | Mitogen-activated protein kinase 14, Myocyte-specific enhancer factor 2A | | Authors: | Chang, C.-I, Xu, B.-E, Akella, R, Cobb, M.H, Goldsmith, E.J. | | Deposit date: | 2002-04-10 | | Release date: | 2002-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of MAP kinase p38 complexed to the docking sites on its nuclear substrate MEF2A and activator MKK3b.
Mol.Cell, 9, 2002
|
|
1LLQ
 
 | | Crystal Structure of Malic Enzyme from Ascaris suum Complexed with Nicotinamide Adenine Dinucleotide | | Descriptor: | NAD-dependent malic enzyme, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Coleman, D.E, Jagannatha, G.S, Goldsmith, E.J, Cook, P.F, Harris, B.G. | | Deposit date: | 2002-04-29 | | Release date: | 2002-05-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the malic enzyme from Ascaris suum complexed with nicotinamide adenine dinucleotide at 2.3 A resolution.
Biochemistry, 41, 2002
|
|
1LEZ
 
 | | CRYSTAL STRUCTURE OF MAP KINASE P38 COMPLEXED TO THE DOCKING SITE ON ITS ACTIVATOR MKK3B | | Descriptor: | MAP kinase kinase 3b, MITOGEN-ACTIVATED PROTEIN KINASE 14 | | Authors: | Chang, C.-I, Xu, B.-E, Akella, R, Cobb, M.H, Goldsmith, E.J. | | Deposit date: | 2002-04-10 | | Release date: | 2002-07-10 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of MAP kinase p38 complexed to the docking sites on its nuclear substrate MEF2A and activator MKK3b.
Mol.Cell, 9, 2002
|
|
1NJJ
 
 | |
1QU4
 
 | | CRYSTAL STRUCTURE OF TRYPANOSOMA BRUCEI ORNITHINE DECARBOXYLASE | | Descriptor: | ORNITHINE DECARBOXYLASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Grishin, N.V, Osterman, A.L, Brooks, H.B, Phillips, M.A, Goldsmith, E.J. | | Deposit date: | 1999-07-06 | | Release date: | 1999-11-17 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray structure of ornithine decarboxylase from Trypanosoma brucei: the native structure and the structure in complex with alpha-difluoromethylornithine.
Biochemistry, 38, 1999
|
|
2TOD
 
 | | ORNITHINE DECARBOXYLASE FROM TRYPANOSOMA BRUCEI K69A MUTANT IN COMPLEX WITH ALPHA-DIFLUOROMETHYLORNITHINE | | Descriptor: | ALPHA-DIFLUOROMETHYLORNITHINE, PROTEIN (ORNITHINE DECARBOXYLASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Grishin, N.V, Osterman, A.L, Brooks, H.B, Phillips, M.A, Goldsmith, E.J. | | Deposit date: | 1999-05-18 | | Release date: | 1999-11-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of ornithine decarboxylase from Trypanosoma brucei: the native structure and the structure in complex with alpha-difluoromethylornithine.
Biochemistry, 38, 1999
|
|
5D9H
 
 | | Crystal structure of SPAK (STK39) dimer in the basal activity state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, STE20/SPS1-related proline-alanine-rich protein kinase, ... | | Authors: | Taylor, C.A, Juang, Y.C, Goldsmith, E.J, Cobb, M.H. | | Deposit date: | 2015-08-18 | | Release date: | 2015-09-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Domain-Swapping Switch Point in Ste20 Protein Kinase SPAK.
Biochemistry, 54, 2015
|
|
1BMK
 
 | | THE COMPLEX STRUCTURE OF THE MAP KINASE P38/SB218655 | | Descriptor: | 4-(FLUOROPHENYL)-1-CYCLOPROPYLMETHYL-5-(2-AMINO-4-PYRIMIDINYL)IMIDAZOLE, PROTEIN (MAP KINASE P38) | | Authors: | Wang, Z, Canagarajah, B, Boehm, J.C, Kassis, S, Cobb, M.H, Young, P.R, Abdel-Meguid, S, Adams, J.L, Goldsmith, E.J. | | Deposit date: | 1998-07-23 | | Release date: | 1999-07-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of inhibitor selectivity in MAP kinases.
Structure, 6, 1998
|
|
1BL7
 
 | | THE COMPLEX STRUCTURE OF THE MAP KINASE P38/SB220025 | | Descriptor: | 4-(4-FLUOROPHENYL)-1-(4-PIPERIDINYL)-5-(2-AMINO-4-PYRIMIDINYL)-IMIDAZOLE, PROTEIN (MAP KINASE P38) | | Authors: | Wang, Z, Canagarajah, B.J, Boehm, J.C, Kassis, S, Cobb, M.H, Young, P.R, Abdel-Meguid, S, Adams, J.L, Goldsmith, E.J. | | Deposit date: | 1998-07-23 | | Release date: | 1999-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of inhibitor selectivity in MAP kinases.
Structure, 6, 1998
|
|
1BL6
 
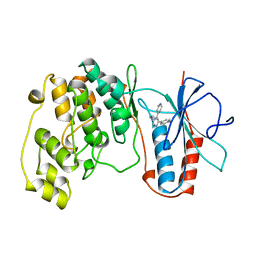 | | THE COMPLEX STRUCTURE OF THE MAP KINASE P38/SB216995 | | Descriptor: | 4-(4-FLUOROPHENYL)-1-CYCLOROPROPYLMETHYL-5-(4-PYRIDYL)-IMIDAZOLE, PROTEIN (MAP KINASE P38) | | Authors: | Wang, Z, Canagarajah, B.J, Boehm, J.C, Kassis, S, Cobb, M.H, Young, P.R, Abdel-Meguid, S, Adams, J.L, Goldsmith, E.J. | | Deposit date: | 1998-07-11 | | Release date: | 1999-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of inhibitor selectivity in MAP kinases.
Structure, 6, 1998
|
|
1A9U
 
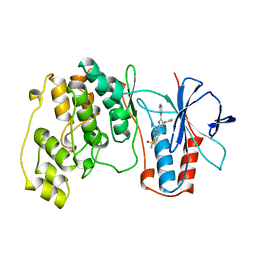 | | THE COMPLEX STRUCTURE OF THE MAP KINASE P38/SB203580 | | Descriptor: | 4-[5-(4-FLUORO-PHENYL)-2-(4-METHANESULFINYL-PHENYL)-3H-IMIDAZOL-4-YL]-PYRIDINE, MAP KINASE P38 | | Authors: | Wang, Z, Canagarajah, B, Boehm, J.C, Kassis, S, Cobb, M.H, Young, P.R, Abdel-Meguid, S, Adams, J.L, Goldsmith, E.J. | | Deposit date: | 1998-04-10 | | Release date: | 1999-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of inhibitor selectivity in MAP kinases.
Structure, 6, 1998
|
|
1PYG
 
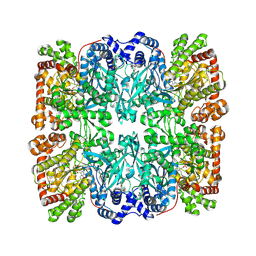 | |
2NVA
 
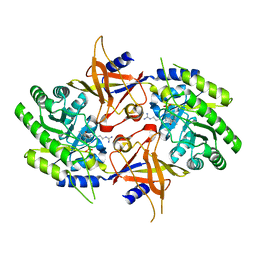 | | The X-ray crystal structure of the Paramecium bursaria Chlorella virus arginine decarboxylase bound to agmatine | | Descriptor: | (4-{[(4-{[AMINO(IMINO)METHYL]AMINO}BUTYL)AMINO]METHYL}-5-HYDROXY-6-METHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, arginine decarboxylase, A207R protein | | Authors: | Shah, R.H, Akella, R, Goldsmith, E, Phillips, M.A. | | Deposit date: | 2006-11-11 | | Release date: | 2007-03-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray Structure of Paramecium bursaria Chlorella Virus Arginine Decarboxylase: Insight into the Structural Basis for Substrate Specificity.
Biochemistry, 46, 2007
|
|
2NV9
 
 | | The X-ray Crystal Structure of the Paramecium bursaria Chlorella virus arginine decarboxylase | | Descriptor: | A207R protein, arginine decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Shah, R.H, Akella, R, Goldsmith, E, Phillips, M.A. | | Deposit date: | 2006-11-11 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | X-ray Structure of Paramecium bursaria Chlorella Virus Arginine Decarboxylase: Insight into the Structural Basis for Substrate Specificity.
Biochemistry, 46, 2007
|
|
4PWN
 
 | | Crystal structure of Active WNK1 kinase | | Descriptor: | PHOSPHATE ION, Serine/threonine-protein kinase WNK1 | | Authors: | Piala, A, Moon, T, Akella, T, He, H, Cobbm, M.H, Goldsmith, E. | | Deposit date: | 2014-03-20 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Chloride Sensing by WNK1 Involves Inhibition of Autophosphorylation.
Sci.Signal., 7, 2014
|
|
4Q2A
 
 | | WNK1: A chloride sensor via autophosphorylation | | Descriptor: | BROMIDE ION, Serine/threonine-protein kinase WNK1 | | Authors: | Piala, A, Moon, T, Akella, R, He, H, Cobb, M.H, Goldsmith, E. | | Deposit date: | 2014-04-07 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Chloride Sensing by WNK1 Involves Inhibition of Autophosphorylation.
Sci.Signal., 7, 2014
|
|
4GPB
 
 | |
3O8A
 
 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with novel Inhibitor Genz667348 | | Descriptor: | Dihydroorotate dehydrogenase homolog, mitochondrial, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Deng, X, Booker, M.L, Phillips, M.A. | | Deposit date: | 2010-08-02 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Novel inhibitors of Plasmodium falciparum dihydroorotate dehydrogenase with anti-malarial activity in the mouse model.
J.Biol.Chem., 285, 2010
|
|
2GPB
 
 | |
3SFK
 
 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM267 | | Descriptor: | 2-(1,1-difluoroethyl)-5-methyl-N-[4-(trifluoromethyl)phenyl][1,2,4]triazolo[1,5-a]pyrimidin-7-amine, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Deng, X, Phillips, M. | | Deposit date: | 2011-06-13 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure-Guided Lead Optimization of Triazolopyrimidine-Ring Substituents Identifies Potent Plasmodium falciparum Dihydroorotate Dehydrogenase Inhibitors with Clinical Candidate Potential.
J.Med.Chem., 54, 2011
|
|
