5H1R
 
 | |
5GLI
 
 | | Human endothelin receptor type-B in the ligand-free form | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Endothelin Receptor Subtype-B, OLEIC ACID, ... | | Authors: | Shihoya, W, Nishizawa, T, Okuta, A, Tani, K, Fujiyoshi, Y, Dohmae, N, Nureki, O, Doi, T. | | Deposit date: | 2016-07-11 | | Release date: | 2016-09-07 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Activation mechanism of endothelin ETB receptor by endothelin-1.
Nature, 537, 2016
|
|
5H1Q
 
 | |
2XZB
 
 | | Pig Gastric H,K-ATPase with bound BeF and SCH28080 | | Descriptor: | POTASSIUM-TRANSPORTING ATPASE ALPHA CHAIN 1, POTASSIUM-TRANSPORTING ATPASE SUBUNIT BETA | | Authors: | Abe, K, Tani, K, Fujiyoshi, Y. | | Deposit date: | 2010-11-24 | | Release date: | 2011-01-26 | | Last modified: | 2020-09-16 | | Method: | ELECTRON CRYSTALLOGRAPHY (7 Å) | | Cite: | Conformational Rearrangement of Gastric H(+),K(+)- ATPase Induced by an Acid Suppressant.
Nat.Commun., 2, 2011
|
|
7F8O
 
 | | Cryo-EM structure of the C-terminal deletion mutant of human PANX1 in a nanodisc | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Pannexin-1 | | Authors: | Kuzuya, M, Hirano, H, Hayashida, K, Watanabe, M, Kobayashi, K, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2021-07-02 | | Release date: | 2022-01-26 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the N terminus and phospholipids.
Sci.Signal., 15, 2022
|
|
7F8J
 
 | | Cryo-EM structure of human pannexin-1 in a nanodisc | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Pannexin-1 | | Authors: | Kuzuya, M, Hirano, H, Hayashida, K, Watanabe, M, Kobayashi, K, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2021-07-02 | | Release date: | 2022-01-26 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the N terminus and phospholipids.
Sci.Signal., 15, 2022
|
|
7F8N
 
 | | Human pannexin-1 showing a conformational change in the N-terminal domain and blocked pore | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Pannexin-1 | | Authors: | Kuzuya, M, Hirano, H, Hayashida, K, Watanabe, M, Kobayashi, K, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2021-07-02 | | Release date: | 2022-01-26 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the N terminus and phospholipids.
Sci.Signal., 15, 2022
|
|
2ZW3
 
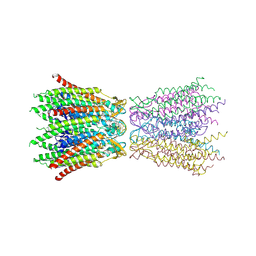 | | Structure of the connexin-26 gap junction channel at 3.5 angstrom resolution | | Descriptor: | Gap junction beta-2 protein | | Authors: | Maeda, S, Nakagawa, S, Suga, M, Yamashita, E, Oshima, A, Fujiyoshi, Y, Tsukihara, T. | | Deposit date: | 2008-12-01 | | Release date: | 2009-04-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the connexin 26 gap junction channel at 3.5 A resolution
Nature, 458, 2009
|
|
2YN9
 
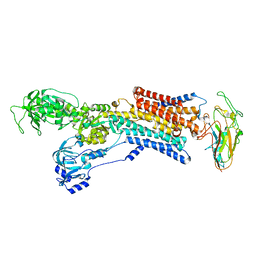 | | Cryo-EM structure of gastric H+,K+-ATPase with bound rubidium | | Descriptor: | POTASSIUM-TRANSPORTING ATPASE ALPHA CHAIN 1, POTASSIUM-TRANSPORTING ATPASE SUBUNIT BETA | | Authors: | Abe, K, Tani, K, Friedrich, T, Fujiyoshi, Y. | | Deposit date: | 2012-10-13 | | Release date: | 2012-11-07 | | Last modified: | 2014-07-16 | | Method: | ELECTRON CRYSTALLOGRAPHY (8 Å) | | Cite: | Cryo-Em Structure of Gastric H+,K+-ATPase with a Single Occupied Cation-Binding Site.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8IF4
 
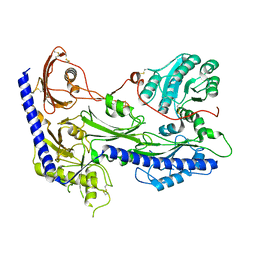 | | Structure of human alpha-2/delta-1 without mirogabalin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Voltage-dependent calcium channel subunit alpha-2/delta-1 | | Authors: | Kozai, D, Numoto, N, Fujiyoshi, Y. | | Deposit date: | 2023-02-17 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Recognition Mechanism of a Novel Gabapentinoid Drug, Mirogabalin, for Recombinant Human alpha 2 delta 1, a Voltage-Gated Calcium Channel Subunit.
J.Mol.Biol., 435, 2023
|
|
8IF3
 
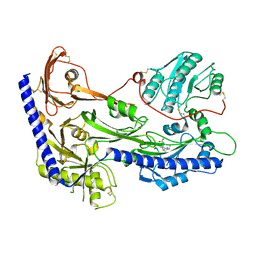 | | Structure of human alpha-2/delta-1 with mirogabalin | | Descriptor: | 2-[(1R,5S,6S)-6-(aminomethyl)-3-ethyl-6-bicyclo[3.2.0]hept-3-enyl]acetic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kozai, D, Numoto, N, Fujiyoshi, Y. | | Deposit date: | 2023-02-17 | | Release date: | 2023-04-05 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Recognition Mechanism of a Novel Gabapentinoid Drug, Mirogabalin, for Recombinant Human alpha 2 delta 1, a Voltage-Gated Calcium Channel Subunit.
J.Mol.Biol., 435, 2023
|
|
6KFG
 
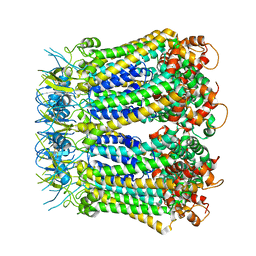 | | Undocked INX-6 hemichannel in detergent | | Descriptor: | Innexin-6 | | Authors: | Burendei, B, Shinozaki, R, Watanabe, M, Terada, T, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2019-07-07 | | Release date: | 2020-02-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of undocked innexin-6 hemichannels in phospholipids.
Sci Adv, 6, 2020
|
|
6KFF
 
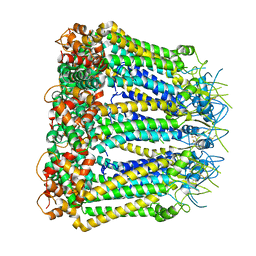 | | Undocked INX-6 hemichannel in a nanodisc | | Descriptor: | Innexin-6 | | Authors: | Burendei, B, Shinozaki, R, Watanabe, M, Terada, T, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2019-07-07 | | Release date: | 2020-02-12 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of undocked innexin-6 hemichannels in phospholipids.
Sci Adv, 6, 2020
|
|
6KFH
 
 | | Undocked hemichannel of an N-terminal deletion mutant of INX-6 in a nanodisc | | Descriptor: | Innexin-6 | | Authors: | Burendei, B, Shinozaki, R, Watanabe, M, Terada, T, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2019-07-07 | | Release date: | 2020-02-12 | | Last modified: | 2020-03-11 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of undocked innexin-6 hemichannels in phospholipids.
Sci Adv, 6, 2020
|
|
5YUA
 
 | | Crystal structure of voltage-gated sodium channel NavAb in high-pH condition | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, CHAPSO, ... | | Authors: | Irie, K, Shimomura, T, Fujiyoshi, Y. | | Deposit date: | 2017-11-21 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.80085564 Å) | | Cite: | Structural insight on the voltage dependence of prokaryotic voltage gated sodium channel NavAb.
FEBS Lett., 2017
|
|
5YUC
 
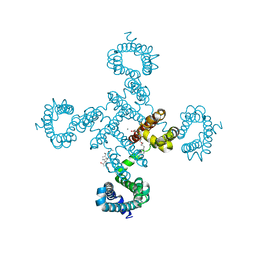 | |
5YUB
 
 | |
6AKG
 
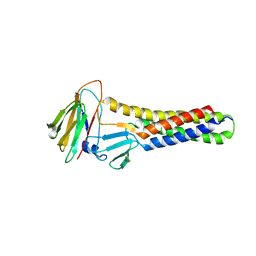 | |
6AKF
 
 | |
6AKE
 
 | |
8GCL
 
 | | Cryo-EM structure of hAQP2 in DDM | | Descriptor: | Aquaporin-2 | | Authors: | Kamegawa, A, Suzuki, S, Nishikawa, K, Numoto, N, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-03-02 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structural analysis of the water channel AQP2 by single-particle cryo-EM.
J.Struct.Biol., 215, 2023
|
|
8HUJ
 
 | | Cryo-EM structure of the J-K-St region of EMCV IRES in complex with eIF4G-HEAT1 and eIF4A | | Descriptor: | Eukaryotic initiation factor 4A-I, Eukaryotic translation initiation factor 4 gamma 1, IRES RNA (J-K-St), ... | | Authors: | Suzuki, H, Fujiyoshi, Y, Imai, S, Shimada, I. | | Deposit date: | 2022-12-24 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Dynamically regulated two-site interaction of viral RNA to capture host translation initiation factor.
Nat Commun, 14, 2023
|
|
8IHI
 
 | | Cryo-EM structure of HCA2-Gi complex with acifran | | Descriptor: | (5~{S})-5-methyl-4-oxidanylidene-5-phenyl-furan-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
8IHF
 
 | | Cryo-EM structure of HCA2-Gi complex with MK6892 | | Descriptor: | 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
8IHK
 
 | | Cryo-EM structure of HCA3-Gi complex with acifran (local) | | Descriptor: | (5~{S})-5-methyl-4-oxidanylidene-5-phenyl-furan-2-carboxylic acid, Soluble cytochrome b562,Hydroxycarboxylic acid receptor 3 | | Authors: | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
