7MSF
 
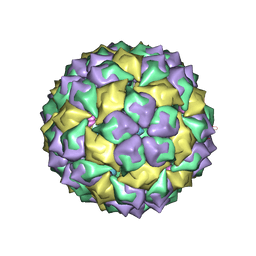 | | MS2 PROTEIN CAPSID/RNA COMPLEX | | Descriptor: | 5'-R(*UP*CP*GP*CP*CP*AP*AP*CP*AP*GP*GP*CP*GP*G)-3', MS2 PROTEIN CAPSID | | Authors: | Rowsell, S, Stonehouse, N.J, Convery, M.A, Adams, C.J, Ellington, A.D, Hirao, I, Peabody, D.S, Stockley, P.G, Phillips, S.E.V. | | Deposit date: | 1998-05-20 | | Release date: | 1998-11-11 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of a series of RNA aptamers complexed to the same protein target.
Nat.Struct.Biol., 5, 1998
|
|
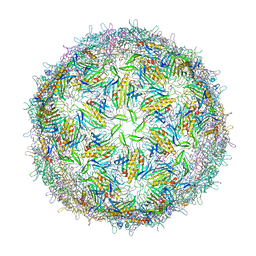
1FRS
 
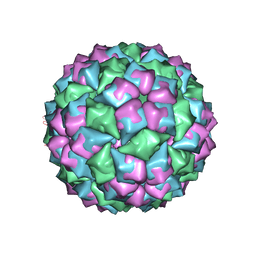 | |
1GAV
 
 | | BACTERIOPHAGE GA PROTEIN CAPSID | | Descriptor: | BACTERIOPHAGE GA PROTEIN CAPSID | | Authors: | Tars, K, Bundule, M, Liljas, L. | | Deposit date: | 1997-01-28 | | Release date: | 1997-09-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The crystal structure of bacteriophage GA and a comparison of bacteriophages belonging to the major groups of Escherichia coli leviviruses.
J.Mol.Biol., 271, 1997
|
|
1CA2
 
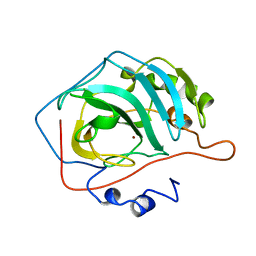 | |
1IKW
 
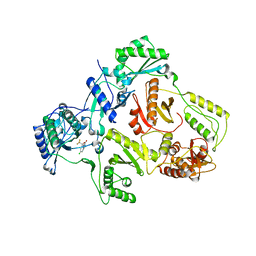 | | Wild Type HIV-1 Reverse Transcriptase in Complex with Efavirenz | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, POL POLYPROTEIN | | Authors: | Lindberg, J, Unge, T. | | Deposit date: | 2001-05-07 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the inhibitory efficacy of efavirenz (DMP-266), MSC194 and PNU142721 towards the HIV-1 RT K103N mutant.
Eur.J.Biochem., 269, 2002
|
|
1IKV
 
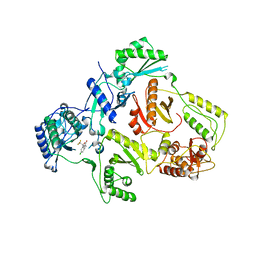 | | K103N Mutant HIV-1 Reverse Transcriptase in Complex with Efivarenz | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, POL POLYPROTEIN | | Authors: | Lindberg, J, Unge, T. | | Deposit date: | 2001-05-07 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the inhibitory efficacy of efavirenz (DMP-266), MSC194 and PNU142721 towards the HIV-1 RT K103N mutant.
Eur.J.Biochem., 269, 2002
|
|
1IKY
 
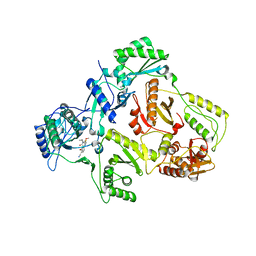 | | HIV-1 Reverse Transcriptase in Complex with the Inhibitor MSC194 | | Descriptor: | 1-[2-(3-ACETYL-2-HYDROXY-6-METHOXY-PHENYL)-CYCLOPROPYL]-3-(5-CYANO-PYRIDIN-2-YL)-THIOUREA, POL POLYPROTEIN | | Authors: | Lindberg, J, Unge, T. | | Deposit date: | 2001-05-07 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the inhibitory efficacy of efavirenz (DMP-266), MSC194 and PNU142721 towards the HIV-1 RT K103N mutant.
Eur.J.Biochem., 269, 2002
|
|
1IKX
 
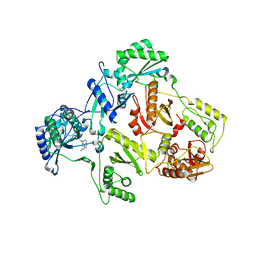 | |
1ZDH
 
 | | MS2 COAT PROTEIN/RNA COMPLEX | | Descriptor: | PROTEIN (BACTERIOPHAGE MS2 COAT PROTEIN), RNA (5'-R(*AP*CP*AP*UP*GP*AP*GP*GP*AP*UP*CP*AP*CP*CP*CP*AP*U P*GP*U)-3') | | Authors: | Valegard, K, Van Den Worm, S, Liljas, L. | | Deposit date: | 1996-09-24 | | Release date: | 1997-04-21 | | Last modified: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The three-dimensional structures of two complexes between recombinant MS2 capsids and RNA operator fragments reveal sequence-specific protein-RNA interactions.
J.Mol.Biol., 270, 1997
|
|
1ZDI
 
 | | RNA BACTERIOPHAGE MS2 COAT PROTEIN/RNA COMPLEX | | Descriptor: | PROTEIN (RNA BACTERIOPHAGE MS2 COAT PROTEIN), RNA (5'-R(*AP*CP*AP*UP*GP*AP*GP*GP*AP*UP*UP*AP*CP*CP*CP*AP*U P*GP*U)-3') | | Authors: | Valegard, K, Van Den Worm, S, Liljas, L. | | Deposit date: | 1996-09-24 | | Release date: | 1997-04-21 | | Last modified: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The three-dimensional structures of two complexes between recombinant MS2 capsids and RNA operator fragments reveal sequence-specific protein-RNA interactions.
J.Mol.Biol., 270, 1997
|
|
1D4H
 
 | | HIV-1 Protease in complex with the inhibitor BEA435 | | Descriptor: | 2,5-DIBENZYLOXY-3,4-DIHYDROXY-HEXANEDIOIC ACID BENZYLAMIDE (2-HYDROXY-INDAN-1-YL)-AMIDE, HIV-1 PROTEASE | | Authors: | Unge, T. | | Deposit date: | 1999-10-04 | | Release date: | 2002-06-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Optimization of P1-P3 groups in symmetric and asymmetric HIV-1 protease inhibitors.
Eur.J.Biochem., 270, 2003
|
|
1D4J
 
 | | HIV-1 protease in complex with the inhibitor MSL370 | | Descriptor: | 2,5-DIBENZYLOXY-3,4-DIHYDROXY-HEXANEDIOIC ACID 2-CHLORO-6-FLUORO-BENZYLAMIDE (2-HYDROXY-INDAN-1- YL)-AMIDE, HIV-1 PROTEASE | | Authors: | Unge, T. | | Deposit date: | 1999-10-04 | | Release date: | 2002-06-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Optimization of P1-P3 groups in symmetric and asymmetric HIV-1 protease inhibitors.
Eur.J.Biochem., 270, 2003
|
|
1EC2
 
 | | HIV-1 protease in complex with the inhibitor BEA428 | | Descriptor: | HIV-1 PROTEASE, N,N-[2,5-O-[DI-4-PYRIDIN-3-YL-BENZYL]-GLUCARYL]-DI-[VALYL-AMIDO-METHANE] | | Authors: | Unge, T. | | Deposit date: | 2000-01-25 | | Release date: | 2002-06-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Optimization of P1-P3 groups in symmetric and asymmetric HIV-1 protease inhibitors
Eur.J.Biochem., 270, 2003
|
|
1EBW
 
 | | HIV-1 protease in complex with the inhibitor BEA322 | | Descriptor: | HIV-1 PROTEASE, N,N-[2,5-O-[DIBENZYL]-GLUCARYL]-DI-[ISOLEUCYL-AMIDO-METHANE] | | Authors: | Unge, T. | | Deposit date: | 2000-01-25 | | Release date: | 2002-06-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Optimization of P1-P3 groups in symmetric and asymmetric HIV-1 protease inhibitors
Eur.J.Biochem., 270, 2003
|
|
1EC3
 
 | | HIV-1 protease in complex with the inhibitor MSA367 | | Descriptor: | HIV-1 PROTEASE, N,N-[2,5-O-DIBENZYL-GLUCARYL]-DI-[VALINYL-AMINOMETHANYL-PYRIDINE] | | Authors: | Unge, T. | | Deposit date: | 2000-01-25 | | Release date: | 2002-06-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Optimization of P1-P3 groups in symmetric and asymmetric HIV-1 protease inhibitors
Eur.J.Biochem., 270, 2003
|
|
1EC1
 
 | | HIV-1 protease in complex with the inhibitor BEA409 | | Descriptor: | HIV-1 PROTEASE, N,N-[2,5-O-[DI-4-THIOPHEN-3-YL-BENZYL]-GLUCARYL]-DI-[VALYL-AMIDO-METHANE] | | Authors: | Unge, T. | | Deposit date: | 2000-01-25 | | Release date: | 2002-06-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Optimization of P1-P3 groups in symmetric and asymmetric HIV-1 protease inhibitors
Eur.J.Biochem., 270, 2003
|
|
1EBY
 
 | | HIV-1 protease in complex with the inhibitor BEA369 | | Descriptor: | HIV-1 PROTEASE, N,N-[2,5-O-DIBENZYL-GLUCARYL]-DI-[1-AMINO-INDAN-2-OL] | | Authors: | Unge, T. | | Deposit date: | 2000-01-25 | | Release date: | 2002-06-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Optimization of P1-P3 groups in symmetric and asymmetric HIV-1 protease inhibitors
Eur.J.Biochem., 270, 2003
|
|
1EBZ
 
 | | HIV-1 protease in complex with the inhibitor BEA388 | | Descriptor: | HIV-1 PROTEASE, [5-(2-HYDROXY-INDAN-1-YLCARBAMOYL)-3,4-DIHYDROXY-2,5-[DIBENZYL-OXY]-PENTANOYL]-VALINYL-AMIDO-METHANE | | Authors: | Unge, T. | | Deposit date: | 2000-01-25 | | Release date: | 2002-06-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Optimization of P1-P3 groups in symmetric and asymmetric HIV-1 protease inhibitors
Eur.J.Biochem., 270, 2003
|
|
1D4I
 
 | | HIV-1 protease in complex with the inhibitor BEA425 | | Descriptor: | 2,5-DIBENZYLOXY-3-HYDROXY-HEXANEDIOIC ACID BIS-[(2-HYDROXY-INDAN-1-YL)-AMIDE], HIV-1 PROTEASE | | Authors: | Unge, T. | | Deposit date: | 1999-10-04 | | Release date: | 2002-06-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Optimization of P1-P3 groups in symmetric and asymmetric HIV-1 protease inhibitors.
Eur.J.Biochem., 270, 2003
|
|
1AZM
 
 | |
2BUK
 
 | | SATELLITE TOBACCO NECROSIS VIRUS | | Descriptor: | CALCIUM ION, COAT PROTEIN | | Authors: | Jones, T.A, Liljas, L. | | Deposit date: | 2005-06-14 | | Release date: | 2005-08-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of Satellite Tobacco Necrosis Virus After Crystallographic Refinement at 2.5 A Resolution.
J.Mol.Biol., 177, 1984
|
|
1CRM
 
 | | STRUCTURE AND FUNCTION OF CARBONIC ANHYDRASES | | Descriptor: | CARBONIC ANHYDRASE I, CHLORIDE ION, HYDROSULFURIC ACID, ... | | Authors: | Yadava, V.S, Kannan, K.K. | | Deposit date: | 1994-03-04 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Function of Carbonic Anhydrases
Biomolecular Structure, Conformation, Function and Evolution, 1, 1981
|
|
1CZM
 
 | |
1BZM
 
 | |
