7PBK
 
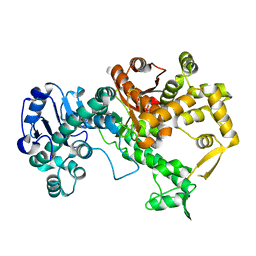 | | Vibriophage phiVC8 family A DNA polymerase (DpoZ), two conformations: thumb-exo open and thumb-exo closed | | Descriptor: | DNA polymerase I | | Authors: | Czernecki, D, Hu, H, Romoli, F, Delarue, M. | | Deposit date: | 2021-08-02 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural dynamics and determinants of 2-aminoadenine specificity in DNA polymerase DpoZ of vibriophage phi VC8.
Nucleic Acids Res., 49, 2021
|
|
3LSV
 
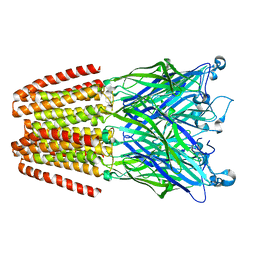 | |
1PYS
 
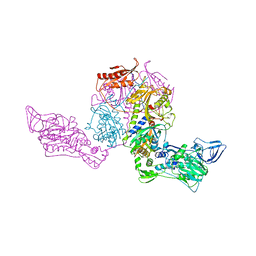 | | PHENYLALANYL-TRNA SYNTHETASE FROM THERMUS THERMOPHILUS | | Descriptor: | MAGNESIUM ION, PHENYLALANYL-TRNA SYNTHETASE | | Authors: | Safro, M, Mosyak, L, Goldgur, Y, Reshetnikova, L, Delarue, M. | | Deposit date: | 1996-11-14 | | Release date: | 1997-11-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of phenylalanyl-tRNA synthetase from Thermus thermophilus.
Nat.Struct.Biol., 2, 1995
|
|
5D4B
 
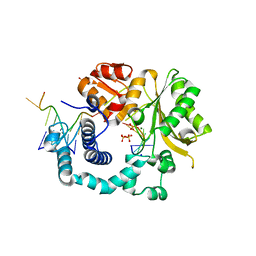 | | Structural Basis for a New Templated Activity by Terminal Deoxynucleotidyl Transferase: Implications for V(D)J Recombination | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*AP*C)-3'), DNA (5'-D(*TP*TP*TP*TP*TP*GP*G)-3'), MAGNESIUM ION, ... | | Authors: | Loc'h, J, Rosario, S, Delarue, M. | | Deposit date: | 2015-08-07 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structural Basis for a New Templated Activity by Terminal Deoxynucleotidyl Transferase: Implications for V(D)J Recombination.
Structure, 24, 2016
|
|
4NPQ
 
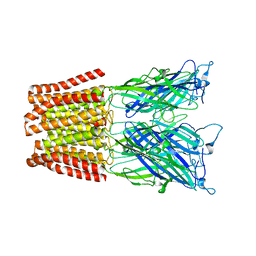 | | The resting-state conformation of the GLIC ligand-gated ion channel | | Descriptor: | Proton-gated ion channel | | Authors: | Sauguet, L, Shahsavar, A, Poitevin, F, Huon, C, Menny, A, Nemecz, A, Haouz, A, Changeux, J.P, Corringer, P.J, Delarue, M. | | Deposit date: | 2013-11-22 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.35 Å) | | Cite: | Crystal structures of a pentameric ligand-gated ion channel provide a mechanism for activation.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4NPP
 
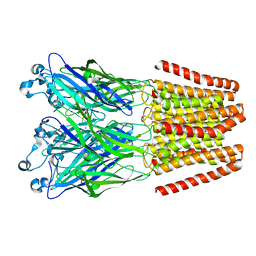 | | The GLIC-His10 wild-type structure in equilibrium between the open and locally-closed (LC) forms | | Descriptor: | NICKEL (II) ION, Proton-gated ion channel | | Authors: | Sauguet, L, Shahsavar, A, Poitevin, F, Huon, C, Menny, A, Nemecz, A, Haouz, A, Changeux, J.P, Corringer, P.J, Delarue, M. | | Deposit date: | 2013-11-22 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Crystal structures of a pentameric ligand-gated ion channel provide a mechanism for activation.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4ZZC
 
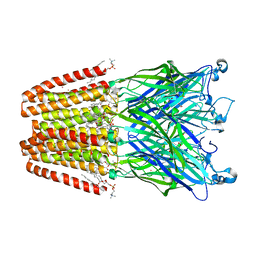 | | The GLIC pentameric Ligand-Gated Ion Channel open form complexed to xenon | | Descriptor: | ACETATE ION, CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Sauguet, L, Fourati, Z, Prange, T, Delarue, M, Colloc'h, N. | | Deposit date: | 2015-05-22 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis for Xenon Inhibition in a Cationic Pentameric Ligand-Gated Ion Channel.
Plos One, 11, 2016
|
|
4ZZB
 
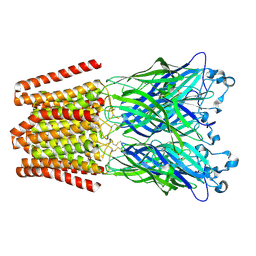 | | The GLIC pentameric Ligand-Gated Ion Channel Locally-closed form complexed to xenon | | Descriptor: | ACETATE ION, CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Sauguet, L, Fourati, Z, Prange, T, Delarue, M, Colloc'h, N. | | Deposit date: | 2015-05-22 | | Release date: | 2016-03-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Basis for Xenon Inhibition in a Cationic Pentameric Ligand-Gated Ion Channel.
Plos One, 11, 2016
|
|
1EQR
 
 | | CRYSTAL STRUCTURE OF FREE ASPARTYL-TRNA SYNTHETASE FROM ESCHERICHIA COLI | | Descriptor: | ASPARTYL-TRNA SYNTHETASE, MAGNESIUM ION | | Authors: | Rees, B, Webster, G, Delarue, M, Boeglin, M, Moras, D. | | Deposit date: | 2000-04-06 | | Release date: | 2000-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Aspartyl tRNA-synthetase from Escherichia coli: flexibility and adaptability to the substrates.
J.Mol.Biol., 299, 2000
|
|
7ODX
 
 | |
7ODY
 
 | |
3P4W
 
 | | Structure of desflurane bound to a pentameric ligand-gated ion channel, GLIC | | Descriptor: | (2S)-2-(difluoromethoxy)-1,1,1,2-tetrafluoroethane, DIUNDECYL PHOSPHATIDYL CHOLINE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Nury, H, Van Renterghem, C, Weng, Y, Tran, A, Baaden, M, Dufresne, V, Changeux, J.P, Sonner, J.M, Delarue, M, Corringer, P.J. | | Deposit date: | 2010-10-07 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray structures of general anaesthetics bound to a pentameric ligand-gated ion channel
Nature, 469, 2011
|
|
3P50
 
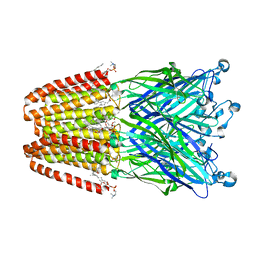 | | Structure of propofol bound to a pentameric ligand-gated ion channel, GLIC | | Descriptor: | 2,6-BIS(1-METHYLETHYL)PHENOL, DIUNDECYL PHOSPHATIDYL CHOLINE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Nury, H, Van Renterghem, C, Weng, Y, Tran, A, Baaden, M, Dufresne, V, Changeux, J.P, Sonner, J.M, Delarue, M, Corringer, P.J. | | Deposit date: | 2010-10-07 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | X-ray structures of general anaesthetics bound to a pentameric ligand-gated ion channel
Nature, 469, 2011
|
|
3IG0
 
 | |
3IFZ
 
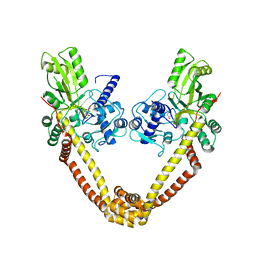 | | crystal structure of the first part of the Mycobacterium tuberculosis DNA gyrase reaction core: the breakage and reunion domain at 2.7 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA gyrase subunit A, SODIUM ION | | Authors: | Piton, J, Aubry, A, Delarue, M, Mayer, C. | | Deposit date: | 2009-07-27 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the quinolone resistance mechanism of Mycobacterium tuberculosis DNA gyrase.
Plos One, 5, 2010
|
|
3IGQ
 
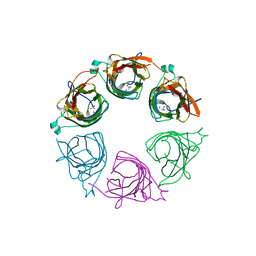 | |
1G51
 
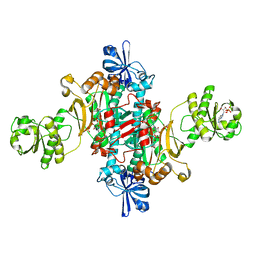 | | ASPARTYL TRNA SYNTHETASE FROM THERMUS THERMOPHILUS AT 2.4 A RESOLUTION | | Descriptor: | ADENOSINE MONOPHOSPHATE, ASPARTYL-ADENOSINE-5'-MONOPHOSPHATE, ASPARTYL-TRNA SYNTHETASE, ... | | Authors: | Poterzsman, A, Delarue, M, Thierry, J.C, Moras, D. | | Deposit date: | 2000-10-30 | | Release date: | 2000-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthesis and recognition of aspartyl-adenylate by Thermus thermophilus aspartyl-tRNA synthetase.
J.Mol.Biol., 244, 1994
|
|
3EAM
 
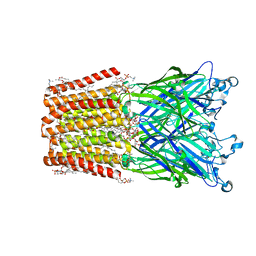 | | An open-pore structure of a bacterial pentameric ligand-gated ion channel | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, DODECYL-BETA-D-MALTOSIDE, Glr4197 protein | | Authors: | Bocquet, N, Nury, H, Baaden, M, Le Poupon, C, Changeux, J.P, Delarue, M, Corringer, P.J. | | Deposit date: | 2008-08-26 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation.
Nature, 457, 2009
|
|
3FI0
 
 | | Crystal Structure Analysis of B. stearothermophilus Tryptophanyl-tRNA Synthetase Complexed with Tryptophan, AMP, and Inorganic Phosphate | | Descriptor: | ADENOSINE MONOPHOSPHATE, PHOSPHATE ION, TRYPTOPHAN, ... | | Authors: | Laowanapiban, P, Kapustina, M, Vonrhein, C, Delarue, M, Koehl, P, Carter Jr, C.W. | | Deposit date: | 2008-12-10 | | Release date: | 2009-02-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Independent saturation of three TrpRS subsites generates a partially assembled state similar to those observed in molecular simulations.
Proc.Natl.Acad.Sci.Usa, 106, 2009
|
|
3FHJ
 
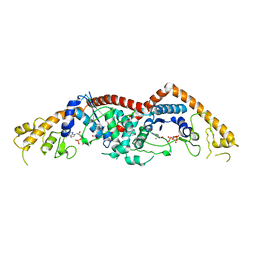 | | Independent saturation of three TrpRS subsites generates a partially-assembled state similar to those observed in molecular simulations | | Descriptor: | ADENOSINE MONOPHOSPHATE, PHOSPHATE ION, TRYPTOPHAN, ... | | Authors: | Laowanapiban, P, Kapustina, M, Vonrhein, C, Delarue, M, Koehl, P, Carter Jr, C.W. | | Deposit date: | 2008-12-09 | | Release date: | 2009-02-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Independent saturation of three TrpRS subsites generates a partially assembled state similar to those observed in molecular simulations.
Proc.Natl.Acad.Sci.Usa, 106, 2009
|
|
3E7F
 
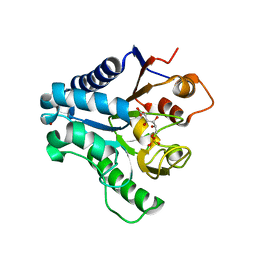 | | Crystal structure of 6-phosphogluconolactonase from Trypanosoma brucei complexed with 6-phosphogluconic acid | | Descriptor: | 6-PHOSPHOGLUCONIC ACID, 6-phosphogluconolactonase, ZINC ION | | Authors: | Poggi, L, Delarue, M, Duclert-Savatier, N, Stoven, V. | | Deposit date: | 2008-08-18 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insights into the enzymatic mechanism of 6-phosphogluconolactonase from Trypanosoma brucei using structural data and molecular dynamics simulation.
J.Mol.Biol., 388, 2009
|
|
3EB9
 
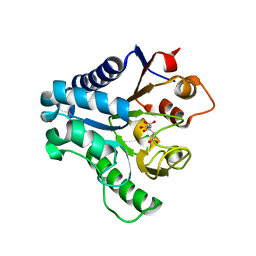 | | Crystal structure of 6-phosphogluconolactonase from trypanosoma brucei complexed with citrate | | Descriptor: | 6-phosphogluconolactonase, CITRATE ANION, ZINC ION | | Authors: | Poggi, L, Delarue, M, Duclert-Savatier, N, Stoven, V. | | Deposit date: | 2008-08-27 | | Release date: | 2009-05-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into the enzymatic mechanism of 6-phosphogluconolactonase from Trypanosoma brucei using structural data and molecular dynamics simulation.
J.Mol.Biol., 388, 2009
|
|
5HCM
 
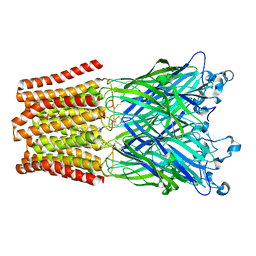 | |
5MUR
 
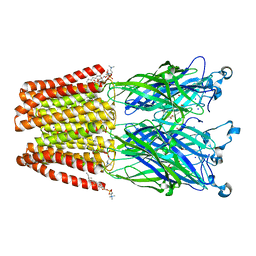 | | X-ray structure of the F14'A mutant of GLIC in complex with propofol | | Descriptor: | 2,6-BIS(1-METHYLETHYL)PHENOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Sauguet, L, Fourati, Z, Delarue, M. | | Deposit date: | 2017-01-13 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis for a Bimodal Allosteric Mechanism of General Anesthetic Modulation in Pentameric Ligand-Gated Ion Channels.
Cell Rep, 23, 2018
|
|
5HCJ
 
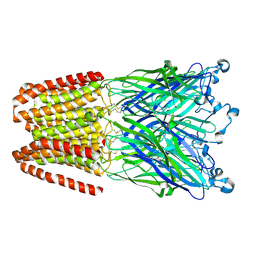 | | Cationic Ligand-Gated Ion Channel | | Descriptor: | CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, Proton-gated ion channel, ... | | Authors: | Shahsavar, A, Sauguet, L, Delarue, M. | | Deposit date: | 2016-01-04 | | Release date: | 2016-04-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Sites of Anesthetic Inhibitory Action on a Cationic Ligand-Gated Ion Channel.
Structure, 24, 2016
|
|
