8IJR
 
 | | The cryo-EM structure of human sphingomyelin synthase-related protein in complex with diacylglycerol/phosphoethanolamine | | Descriptor: | (2S)-1-(hexadecanoyloxy)-3-hydroxypropan-2-yl (11Z)-octadec-11-enoate, PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER, Sphingomyelin synthase-related protein 1 | | Authors: | Hu, K, Zhang, Q, Chen, Y, Yao, D, Zhou, L, Cao, Y. | | Deposit date: | 2023-02-28 | | Release date: | 2024-02-28 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Cryo-EM structure of human sphingomyelin synthase and its mechanistic implications for sphingomyelin synthesis.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8IJQ
 
 | | The cryo-EM structure of human sphingomyelin synthase-related protein in complex with ceramide | | Descriptor: | N-((E,2S,3R)-1,3-DIHYDROXYOCTADEC-4-EN-2-YL)PALMITAMIDE, Sphingomyelin synthase-related protein 1 | | Authors: | Hu, K, Zhang, Q, Chen, Y, Yao, D, Zhou, L, Cao, Y. | | Deposit date: | 2023-02-27 | | Release date: | 2024-02-28 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Cryo-EM structure of human sphingomyelin synthase and its mechanistic implications for sphingomyelin synthesis.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7VQK
 
 | |
7Y53
 
 | | The cryo-EM structure of human ERAD retro-translocation complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Derlin-1, Transitional endoplasmic reticulum ATPase | | Authors: | Cao, Y, Rao, B, Wang, Q, Yao, D, Xia, Y, Li, W, Li, S, Shen, Y. | | Deposit date: | 2022-06-16 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | The cryo-EM structure of the human ERAD retrotranslocation complex.
Sci Adv, 9, 2023
|
|
7Y59
 
 | | The cryo-EM structure of human ERAD retro-translocation complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Derlin-1, ... | | Authors: | Cao, Y, Rao, B, Wang, Q, Yao, D, Xia, Y, Li, W, Li, S, Shen, Y. | | Deposit date: | 2022-06-16 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (4.51 Å) | | Cite: | The cryo-EM structure of the human ERAD retrotranslocation complex.
Sci Adv, 9, 2023
|
|
7Y4W
 
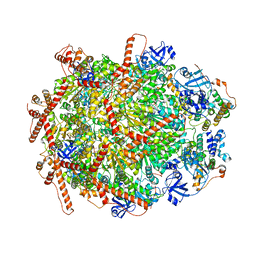 | | The cryo-EM structure of human ERAD retro-translocation complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Derlin-1, Transitional endoplasmic reticulum ATPase | | Authors: | Cao, Y, Rao, B, Wang, Q, Yao, D, Xia, Y, Li, W, Li, S, Shen, Y. | | Deposit date: | 2022-06-16 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | The cryo-EM structure of the human ERAD retrotranslocation complex.
Sci Adv, 9, 2023
|
|
6JYM
 
 | |
6KIZ
 
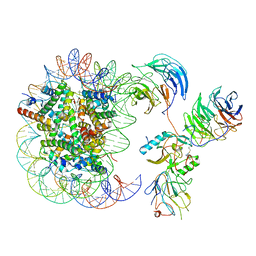 | | Cryo-EM structure of human MLL1-NCP complex, binding mode2 | | Descriptor: | DNA (145-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Huang, J, Xue, H, Yao, T. | | Deposit date: | 2019-07-20 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural basis of nucleosome recognition and modification by MLL methyltransferases.
Nature, 573, 2019
|
|
6KIV
 
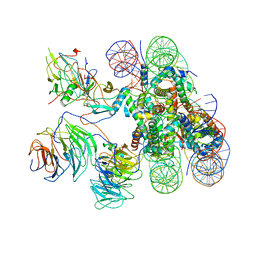 | | Cryo-EM structure of human MLL1-ubNCP complex (4.0 angstrom) | | Descriptor: | DNA (145-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Huang, J, Xue, H, Yao, T. | | Deposit date: | 2019-07-20 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of nucleosome recognition and modification by MLL methyltransferases.
Nature, 573, 2019
|
|
6KIU
 
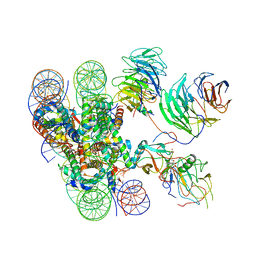 | | Cryo-EM structure of human MLL1-ubNCP complex (3.2 angstrom) | | Descriptor: | DNA (145-MER), GLUTAMINE, Histone H2A, ... | | Authors: | Huang, J, Xue, H, Yao, T. | | Deposit date: | 2019-07-20 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of nucleosome recognition and modification by MLL methyltransferases.
Nature, 573, 2019
|
|
6KIW
 
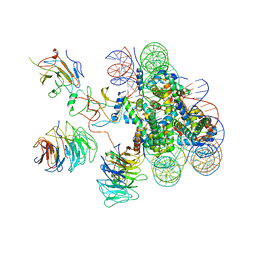 | | Cryo-EM structure of human MLL3-ubNCP complex (4.0 angstrom) | | Descriptor: | DNA (144-MER), DNA (145-MER), Histone H2A, ... | | Authors: | Huang, J, Xue, H, Yao, T. | | Deposit date: | 2019-07-20 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of nucleosome recognition and modification by MLL methyltransferases.
Nature, 573, 2019
|
|
6KI0
 
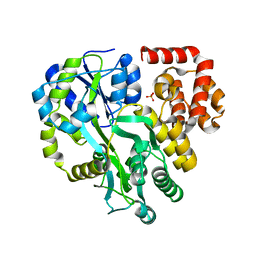 | | Crystal Structure of Human ASC-CARD | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Apoptosis-associated speck-like protein containing a CARD, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Xu, Z.H, Jin, T.C. | | Deposit date: | 2019-07-16 | | Release date: | 2020-07-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Homotypic CARD-CARD interaction is critical for the activation of NLRP1 inflammasome.
Cell Death Dis, 12, 2021
|
|
6KIX
 
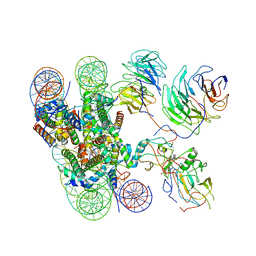 | | Cryo-EM structure of human MLL1-NCP complex, binding mode1 | | Descriptor: | DNA (145-MER), GLUTAMINE, Histone H2A, ... | | Authors: | Huang, J, Xue, H, Yao, T. | | Deposit date: | 2019-07-20 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of nucleosome recognition and modification by MLL methyltransferases.
Nature, 573, 2019
|
|
5Y36
 
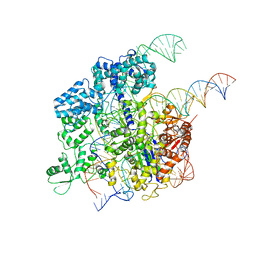 | | Cryo-EM structure of SpCas9-sgRNA-DNA ternary complex | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, MAGNESIUM ION, complementary DNA strand, ... | | Authors: | Huang, Q, Li, G, Huai, C. | | Deposit date: | 2017-07-27 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Structural insights into DNA cleavage activation of CRISPR-Cas9 system
Nat Commun, 8, 2017
|
|
7C7A
 
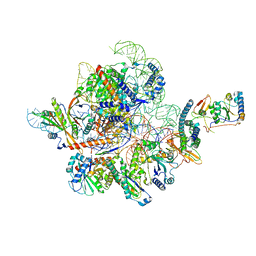 | | Cryo-EM structure of yeast Ribonuclease MRP with substrate ITS1 | | Descriptor: | Internal Transcribed Spacer 1, MAGNESIUM ION, RNases MRP/P 32.9 kDa subunit, ... | | Authors: | Lan, P, Wu, J, Lei, M. | | Deposit date: | 2020-05-24 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insight into precursor ribosomal RNA processing by ribonuclease MRP.
Science, 369, 2020
|
|
7C79
 
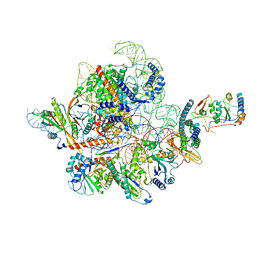 | | Cryo-EM structure of yeast Ribonuclease MRP | | Descriptor: | MAGNESIUM ION, RNases MRP/P 32.9 kDa subunit, Ribonuclease MRP RNA subunit NME1, ... | | Authors: | Lan, P, Wu, J, Lei, M. | | Deposit date: | 2020-05-24 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural insight into precursor ribosomal RNA processing by ribonuclease MRP.
Science, 369, 2020
|
|
7D58
 
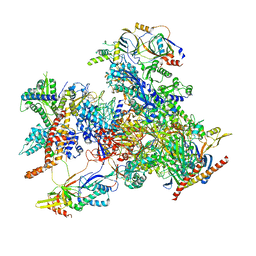 | | cryo-EM structure of human RNA polymerase III in elongating state | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Wang, Q, Wan, F, Lan, P, Wu, J, Lei, M. | | Deposit date: | 2020-09-25 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into transcriptional regulation of human RNA polymerase III.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7D59
 
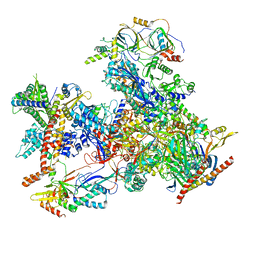 | | cryo-EM structure of human RNA polymerase III in apo state | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Wang, Q, Wan, F, Lan, P, Wu, J, Lei, M. | | Deposit date: | 2020-09-25 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into transcriptional regulation of human RNA polymerase III.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7WTD
 
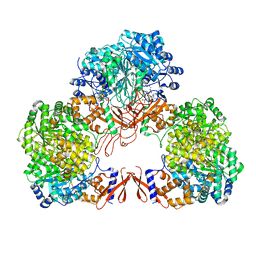 | | Cryo-EM structure of human pyruvate carboxylase with acetyl-CoA in the intermediate state 1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, COENZYME A, Pyruvate carboxylase, ... | | Authors: | Chai, P, Lan, P, Wu, J, Lei, M. | | Deposit date: | 2022-02-04 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mechanistic insight into allosteric activation of human pyruvate carboxylase by acetyl-CoA.
Mol.Cell, 82, 2022
|
|
7WTE
 
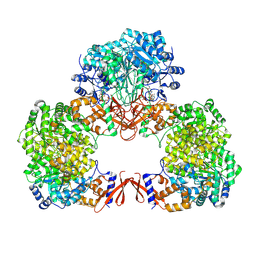 | | Cryo-EM structure of human pyruvate carboxylase with acetyl-CoA in the intermediate state 2 | | Descriptor: | ACETYL COENZYME *A, ADENOSINE-5'-TRIPHOSPHATE, Pyruvate carboxylase, ... | | Authors: | Chai, P, Lan, P, Wu, J, Lei, M. | | Deposit date: | 2022-02-04 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanistic insight into allosteric activation of human pyruvate carboxylase by acetyl-CoA.
Mol.Cell, 82, 2022
|
|
7WTA
 
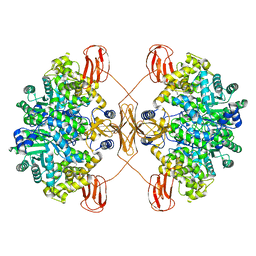 | | Cryo-EM structure of human pyruvate carboxylase in apo state | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Pyruvate carboxylase, mitochondrial | | Authors: | Chai, P, Lan, P, Wu, J, Lei, M. | | Deposit date: | 2022-02-04 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mechanistic insight into allosteric activation of human pyruvate carboxylase by acetyl-CoA.
Mol.Cell, 82, 2022
|
|
7WTC
 
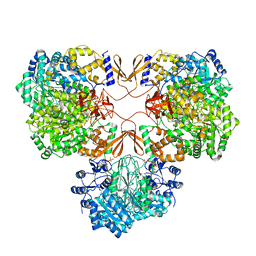 | | Cryo-EM structure of human pyruvate carboxylase with acetyl-CoA in the ground state | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ACETYL COENZYME *A, Pyruvate carboxylase, ... | | Authors: | Chai, P, Lan, P, Wu, J, Lei, M. | | Deposit date: | 2022-02-04 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Mechanistic insight into allosteric activation of human pyruvate carboxylase by acetyl-CoA.
Mol.Cell, 82, 2022
|
|
7WTB
 
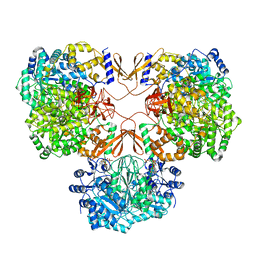 | | Cryo-EM structure of human pyruvate carboxylase with acetyl-CoA | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ACETYL COENZYME *A, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Chai, P, Lan, P, Wu, J, Lei, M. | | Deposit date: | 2022-02-04 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanistic insight into allosteric activation of human pyruvate carboxylase by acetyl-CoA.
Mol.Cell, 82, 2022
|
|
7EA8
 
 | |
7EA5
 
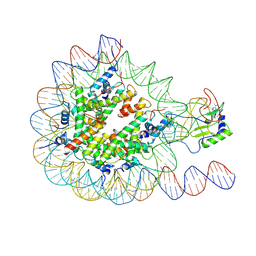 | |
