7FCI
 
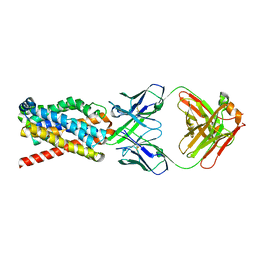 | | human NTCP in complex with YN69083 Fab | | Descriptor: | Fab Heavy chain, Fab Light chain, Sodium/bile acid cotransporter | | Authors: | Park, J.H, Iwamoto, M, Yun, J.H, Uchikubo-Kamo, T, Son, D, Jin, Z, Yoshida, H, Ohki, M, Ishimoto, N, Mizutani, K, Oshima, M, Muramatsu, M, Wakita, T, Shirouzu, M, Liu, K, Uemura, T, Nomura, N, Iwata, S, Watashi, K, Tame, J.R.H, Nishizawa, T, Lee, W, Park, S.Y. | | Deposit date: | 2021-07-14 | | Release date: | 2022-05-25 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into the HBV receptor and bile acid transporter NTCP.
Nature, 606, 2022
|
|
1ZRJ
 
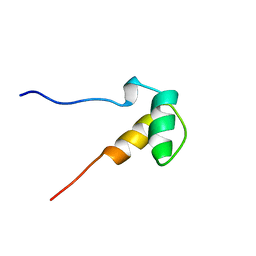 | | Solution structure of the SAP domain of human E1B-55kDa-associated protein 5 isoform c | | Descriptor: | E1B-55kDa-associated protein 5 isoform c | | Authors: | Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-19 | | Release date: | 2005-11-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SAP domain of human E1B-55kDa-associated protein 5 isoform c
To be Published
|
|
6AJ4
 
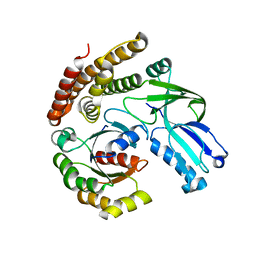 | |
6AJL
 
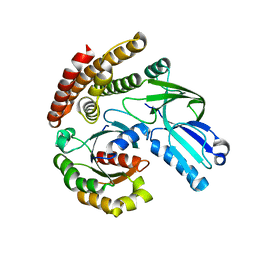 | | DOCK7 mutant I1836Y complexed with Cdc42 | | Descriptor: | Cell division control protein 42 homolog, Dedicator of cytokinesis protein 7 | | Authors: | Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2018-08-28 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Structural Basis for the Dual Substrate Specificity of DOCK7 Guanine Nucleotide Exchange Factor.
Structure, 27, 2019
|
|
5ZQQ
 
 | | Tankyrase-2 in complex with compound 52 | | Descriptor: | 1-methyl-1'-(4-oxo-3,4,5,6,7,8-hexahydroquinazolin-2-yl)spiro[indole-3,4'-piperidin]-2(1H)-one, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Yoshimoto, N, Tsumura, T, Okue, M, Shirouzu, M, Seimiya, H, Umehara, T. | | Deposit date: | 2018-04-19 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Discovery of Novel Spiroindoline Derivatives as Selective Tankyrase Inhibitors.
J. Med. Chem., 62, 2019
|
|
5ZQO
 
 | | Tankyrase-2 in complex with compound 1a | | Descriptor: | 2-[4-(2-methoxyphenyl)piperazin-1-yl]-5,6,7,8-tetrahydroquinazolin-4(3H)-one, GLYCEROL, SULFATE ION, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Yoshimoto, N, Tsumura, T, Okue, M, Shirouzu, M, Seimiya, H, Umehara, T. | | Deposit date: | 2018-04-19 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Discovery of Novel Spiroindoline Derivatives as Selective Tankyrase Inhibitors.
J. Med. Chem., 62, 2019
|
|
5ZSU
 
 | | Structure of the human homo-hexameric LRRC8A channel at 4.25 Angstroms | | Descriptor: | Volume-regulated anion channel subunit LRRC8A | | Authors: | Kasuya, G, Nakane, T, Yokoyama, T, Shirouzu, M, Ishitani, R, Nureki, O. | | Deposit date: | 2018-04-29 | | Release date: | 2018-08-15 | | Last modified: | 2018-09-26 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Cryo-EM structures of the human volume-regulated anion channel LRRC8.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5ZQP
 
 | | Tankyrase-2 in complex with compound 12 | | Descriptor: | 1'-(4-oxo-3,4,5,6,7,8-hexahydroquinazolin-2-yl)-2H-spiro[1-benzofuran-3,4'-piperidin]-2-one, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Yoshimoto, N, Tsumura, T, Okue, M, Shirouzu, M, Seimiya, H, Umehara, T. | | Deposit date: | 2018-04-19 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Discovery of Novel Spiroindoline Derivatives as Selective Tankyrase Inhibitors.
J. Med. Chem., 62, 2019
|
|
5ZQR
 
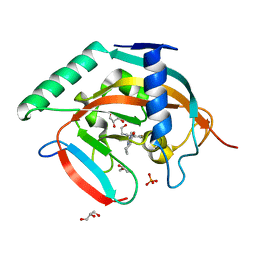 | | Tankyrase-2 in complex with compound 40c | | Descriptor: | 2-[4,6-difluoro-1-(2-hydroxyethyl)-1,2-dihydro-1'H-spiro[indole-3,4'-piperidin]-1'-yl]-5,6,7,8-tetrahydroquinazolin-4(3H)-one, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Yoshimoto, N, Tsumura, T, Okue, M, Shirouzu, M, Seimiya, H, Umehara, T. | | Deposit date: | 2018-04-19 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Novel Spiroindoline Derivatives as Selective Tankyrase Inhibitors.
J. Med. Chem., 62, 2019
|
|
6A84
 
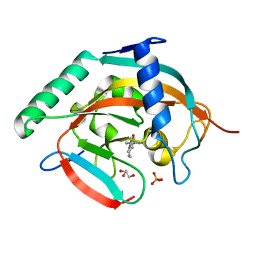 | | Tankyrase-2 in complex with compound 15d | | Descriptor: | 2-(4-chloro-1,2-dihydro-1'H-spiro[indole-3,4'-piperidin]-1'-yl)-5,6,7,8-tetrahydroquinazolin-4(3H)-one, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Yoshimoto, N, Tsumura, T, Okue, M, Shirouzu, M, Seimiya, H, Umehara, T. | | Deposit date: | 2018-07-06 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery of Novel Spiroindoline Derivatives as Selective Tankyrase Inhibitors.
J. Med. Chem., 62, 2019
|
|
8ZJI
 
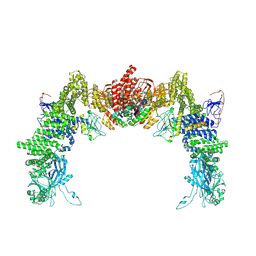 | | Structure of DOCK5/ELMO1/Rac1 core (RhoG/DOCK5/ELMO1/Rac1 dataset, class 1) | | Descriptor: | Dedicator of cytokinesis protein 5, Engulfment and cell motility protein 1, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Kukimoto-Niino, M, Katsura, K, Ishizuka-Katsura, Y, Mishima-Tsumagari, C, Yonemochi, M, Inoue, M, Nakagawa, R, Kaushik, R, Zhang, K.Y.J, Shirouzu, M. | | Deposit date: | 2024-05-15 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | RhoG facilitates a conformational transition in the guanine nucleotide exchange factor complex DOCK5/ELMO1 to an open state.
J.Biol.Chem., 300, 2024
|
|
8ZJK
 
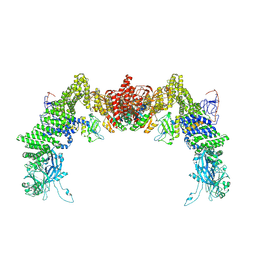 | | Structure of DOCK5/ELMO1/Rac1 core (RhoG/DOCK5/ELMO1/Rac1 dataset, class 3) | | Descriptor: | Dedicator of cytokinesis protein 5, Engulfment and cell motility protein 1, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Kukimoto-Niino, M, Katsura, K, Ishizuka-Katsura, Y, Mishima-Tsumagari, C, Yonemochi, M, Inoue, M, Nakagawa, R, Kaushik, R, Zhang, K.Y.J, Shirouzu, M. | | Deposit date: | 2024-05-15 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | RhoG facilitates a conformational transition in the guanine nucleotide exchange factor complex DOCK5/ELMO1 to an open state.
J.Biol.Chem., 300, 2024
|
|
8ZJM
 
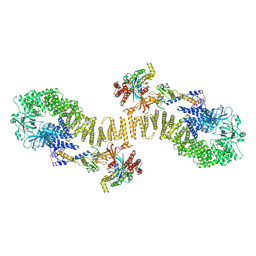 | | Structure of DOCK5/ELMO1/Rac1 core (RhoG/DOCK5/ELMO1/Rac1 dataset, class 5) | | Descriptor: | Dedicator of cytokinesis protein 5, Engulfment and cell motility protein 1, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Kukimoto-Niino, M, Katsura, K, Ishizuka-Katsura, Y, Mishima-Tsumagari, C, Yonemochi, M, Inoue, M, Nakagawa, R, Kaushik, R, Zhang, K.Y.J, Shirouzu, M. | | Deposit date: | 2024-05-15 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.52 Å) | | Cite: | RhoG facilitates a conformational transition in the guanine nucleotide exchange factor complex DOCK5/ELMO1 to an open state.
J.Biol.Chem., 300, 2024
|
|
8ZJL
 
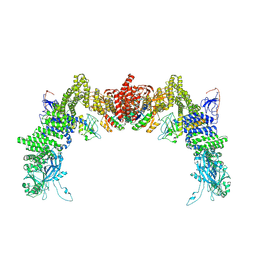 | | Structure of DOCK5/ELMO1/Rac1 core (RhoG/DOCK5/ELMO1/Rac1 dataset, class 4) | | Descriptor: | Dedicator of cytokinesis protein 5, Engulfment and cell motility protein 1, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Kukimoto-Niino, M, Katsura, K, Ishizuka-Katsura, Y, Mishima-Tsumagari, C, Yonemochi, M, Inoue, M, Nakagawa, R, Kaushik, R, Zhang, K.Y.J, Shirouzu, M. | | Deposit date: | 2024-05-15 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.31 Å) | | Cite: | RhoG facilitates a conformational transition in the guanine nucleotide exchange factor complex DOCK5/ELMO1 to an open state.
J.Biol.Chem., 300, 2024
|
|
8ZJJ
 
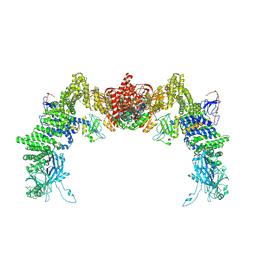 | | Structure of DOCK5/ELMO1/Rac1 core (RhoG/DOCK5/ELMO1/Rac1 dataset, class 2) | | Descriptor: | Dedicator of cytokinesis protein 5, Engulfment and cell motility protein 1, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Kukimoto-Niino, M, Katsura, K, Ishizuka-Katsura, Y, Mishima-Tsumagari, C, Yonemochi, M, Inoue, M, Nakagawa, R, Kaushik, R, Zhang, K.Y.J, Shirouzu, M. | | Deposit date: | 2024-05-15 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | RhoG facilitates a conformational transition in the guanine nucleotide exchange factor complex DOCK5/ELMO1 to an open state.
J.Biol.Chem., 300, 2024
|
|
8XM7
 
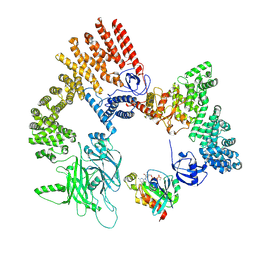 | | Cryo-EM structure of the RhoG/DOCK5/ELMO1/Rac1 complex: RhoG/DOCK5/ELMO1 focused map | | Descriptor: | Dedicator of cytokinesis protein 5, Engulfment and cell motility protein 1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Kukimoto-Niino, M, Katsura, K, Ishizuka-Katsura, Y, Mishima-Tsumagari, C, Yonemochi, M, Inoue, M, Nakagawa, R, Kaushik, R, Zhang, K.Y.J, Shirouzu, M. | | Deposit date: | 2023-12-27 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.91 Å) | | Cite: | RhoG facilitates a conformational transition in the guanine nucleotide exchange factor complex DOCK5/ELMO1 to an open state.
J.Biol.Chem., 300, 2024
|
|
8ZJ2
 
 | | Cryo-EM structure of the RhoG/DOCK5/ELMO1/Rac1 complex | | Descriptor: | Dedicator of cytokinesis protein 5, Engulfment and cell motility protein 1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Kukimoto-Niino, M, Katsura, K, Ishizuka-Katsura, Y, Mishima-Tsumagari, C, Yonemochi, M, Inoue, M, Nakagawa, R, Kaushik, R, Zhang, K.Y.J, Shirouzu, M. | | Deposit date: | 2024-05-14 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.66 Å) | | Cite: | RhoG facilitates a conformational transition in the guanine nucleotide exchange factor complex DOCK5/ELMO1 to an open state.
J.Biol.Chem., 300, 2024
|
|
5KNC
 
 | | Crystal structure of the 3 ADP-bound V1 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Suzuki, K, Mizutani, K, Maruyama, S, Shimono, K, Imai, F.L, Muneyuki, E, Kakinuma, Y, Ishizuka-Katsura, Y, Shirouzu, M, Yokoyama, S, Yamato, I, Murata, T. | | Deposit date: | 2016-06-28 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.015 Å) | | Cite: | Crystal structures of the ATP-binding and ADP-release dwells of the V1 rotary motor
Nat Commun, 7, 2016
|
|
5KNB
 
 | | Crystal structure of the 2 ADP-bound V1 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Suzuki, K, Mizutani, K, Maruyama, S, Shimono, K, Imai, F.L, Muneyuki, E, Kakinuma, Y, Ishizuka-Katsura, Y, Shirouzu, M, Yokoyama, S, Yamato, I, Murata, T. | | Deposit date: | 2016-06-28 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.251 Å) | | Cite: | Crystal structures of the ATP-binding and ADP-release dwells of the V1 rotary motor
Nat Commun, 7, 2016
|
|
1RRB
 
 | | THE RAS-BINDING DOMAIN OF RAF-1 FROM RAT, NMR, 1 STRUCTURE | | Descriptor: | RAF PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE | | Authors: | Terada, T, Ito, Y, Shirouzu, M, Tateno, M, Hashimoto, K, Kigawa, T, Ebisuzaki, T, Takio, K, Shibata, T, Yokoyama, S, Smith, B.O, Laue, E.D, Cooper, J.A, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1998-03-26 | | Release date: | 1999-03-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance and molecular dynamics studies on the interactions of the Ras-binding domain of Raf-1 with wild-type and mutant Ras proteins.
J.Mol.Biol., 286, 1999
|
|
1J1H
 
 | | Solution structure of a tmRNA-binding protein, SmpB, from Thermus thermophilus | | Descriptor: | Small Protein B | | Authors: | Someya, T, Nameki, N, Hosoi, H, Suzuki, S, Hatanaka, H, Fujii, M, Terada, T, Shirouzu, M, Inoue, Y, Shibata, T, Kuramitsu, S, Yokoyama, S, Kawai, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-12-04 | | Release date: | 2003-02-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a tmRNA-binding protein, SmpB, from Thermus thermophilus
FEBS Lett., 535, 2003
|
|
1J03
 
 | | Solution structure of a putative steroid-binding protein from Arabidopsis | | Descriptor: | putative steroid binding protein | | Authors: | Suzuki, S, Hatanaka, H, Kigawa, T, Terada, T, Shirouzu, M, Seki, M, Shinozaki, K, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-10-29 | | Release date: | 2003-12-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an Arabidopsis homologue of the mammalian membrane-associated progesterone receptor
To be Published
|
|
1ZPW
 
 | |
1Z54
 
 | | Crystal structure of a hypothetical protein TT1821 from Thermus thermophilus | | Descriptor: | GLYCEROL, probable thioesterase | | Authors: | Ihsanawati, Kaminishi, T, Murayama, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-03-17 | | Release date: | 2005-09-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a hypothetical protein TT1821 from Thermus thermophilus
TO BE PUBLISHED
|
|
2MGZ
 
 | | Solution structure of RBFOX family ASD-1 RRM and SUP-12 RRM in ternary complex with RNA | | Descriptor: | Protein ASD-1, isoform a, Protein SUP-12, ... | | Authors: | Takahashi, M, Kuwasako, K, Unzai, S, Tsuda, K, Yoshikawa, S, He, F, Kobayashi, N, Guntert, P, Shirouzu, M, Ito, T, Tanaka, A, Yokoyama, S, Hagiwara, M, Kuroyanagi, H, Muto, Y. | | Deposit date: | 2013-11-12 | | Release date: | 2014-08-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | RBFOX and SUP-12 sandwich a G base to cooperatively regulate tissue-specific splicing
Nat.Struct.Mol.Biol., 21, 2014
|
|
