4WBQ
 
 | | Crystal structure of the exonuclease domain of QIP (QDE-2 interacting protein) solved by native-SAD phasing. | | Descriptor: | CALCIUM ION, QDE-2-interacting protein | | Authors: | Boland, A, Weinert, T, Weichenrieder, O, Wang, M. | | Deposit date: | 2014-09-03 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
4WBX
 
 | | Conserved hypothetical protein PF1771 from Pyrococcus furiosus solved by sulfur SAD using Swiss Light Source data | | Descriptor: | 2-keto acid:ferredoxin oxidoreductase subunit alpha | | Authors: | Weinert, T, Waltersperger, S, Olieric, V, Panepucci, E, Chen, L, Rose, J.P, Wang, M, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2014-09-04 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
4WAU
 
 | | Crystal structure of CENP-M solved by native-SAD phasing | | Descriptor: | Centromere protein M | | Authors: | Weinert, T, Basilico, F, Cecatiello, V, Pasqualato, S, Wang, M. | | Deposit date: | 2014-09-01 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
4WAB
 
 | | Crystal structure of mPGES1 solved by native-SAD phasing | | Descriptor: | 2-[[2,6-bis(chloranyl)-3-[(2,2-dimethylpropanoylamino)methyl]phenyl]amino]-1-methyl-6-(2-methyl-2-oxidanyl-propoxy)-N-[2,2,2-tris(fluoranyl)ethyl]benzimidazole-5-carboxamide, GLUTATHIONE, Prostaglandin E synthase,Leukotriene C4 synthase | | Authors: | Weinert, T, Li, D, Howe, N, Caffrey, M, Wang, M. | | Deposit date: | 2014-08-29 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
4BPM
 
 | | Crystal structure of a human integral membrane enzyme | | Descriptor: | 2-[[2,6-bis(chloranyl)-3-[(2,2-dimethylpropanoylamino)methyl]phenyl]amino]-1-methyl-6-(2-methyl-2-oxidanyl-propoxy)-N-[2,2,2-tris(fluoranyl)ethyl]benzimidazole-5-carboxamide, GLUTATHIONE, PROSTAGLANDIN E SYNTHASE, ... | | Authors: | Li, D, Wang, M, Olieric, V, Caffrey, M. | | Deposit date: | 2013-05-27 | | Release date: | 2014-04-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystallizing Membrane Proteins in the Lipidic Mesophase. Experience with Human Prostaglandin E2 Synthase 1 and an Evolving Strategy.
Cryst.Growth Des., 14, 2014
|
|
5HES
 
 | | Human leucine zipper- and sterile alpha motif-containing kinase (ZAK, MLT, HCCS-4, MRK, AZK, MLTK) in complex with vemurafenib | | Descriptor: | 1,2-ETHANEDIOL, Mitogen-activated protein kinase kinase kinase MLT, N-(3-{[5-(4-chlorophenyl)-1H-pyrrolo[2,3-b]pyridin-3-yl]carbonyl}-2,4-difluorophenyl)propane-1-sulfonamide | | Authors: | Mathea, S, Salah, E, Abdul Azeez, K.R, Tallant, C, Szklarz, M, Chaikuad, A, Shrestha, B, Sorrell, F.J, Elkins, J.M, Shrestha, L, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S. | | Deposit date: | 2016-01-06 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structure of the Human Protein Kinase ZAK in Complex with Vemurafenib.
Acs Chem.Biol., 11, 2016
|
|
8CNI
 
 | | PHT1 in the outward facing conformation, bound to Sb27 | | Descriptor: | Solute carrier family 15 member 4, Sybody 27 | | Authors: | Custodio, T, Killer, M, Loew, C. | | Deposit date: | 2023-02-23 | | Release date: | 2023-09-27 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Molecular basis of TASL recruitment by the peptide/histidine transporter 1, PHT1.
Nat Commun, 14, 2023
|
|
1E2J
 
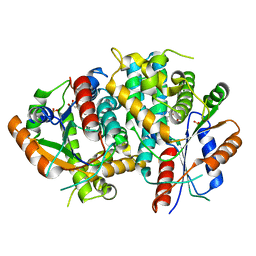 | | The nucleoside binding site of Herpes simplex type 1 thymidine kinase analyzed by X-ray crystallography | | Descriptor: | SULFATE ION, THYMIDINE, THYMIDINE KINASE | | Authors: | Vogt, J, Scapozza, L, Schulz, G.E. | | Deposit date: | 2000-05-23 | | Release date: | 2000-11-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nucleoside Binding Site of Herpes Simplex Type 1 Thymidine Kinase Analyzed by X-Ray Crystallography
Proteins: Struct.,Funct., Genet., 41, 2000
|
|
1E2H
 
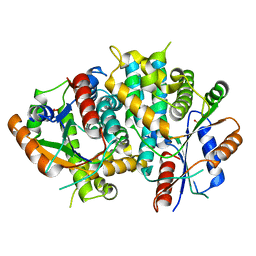 | | The nucleoside binding site of Herpes simplex type 1 thymidine kinase analyzed by X-ray crystallography | | Descriptor: | SULFATE ION, THYMIDINE KINASE | | Authors: | Vogt, J, Scapozza, L, Schulz, G.E. | | Deposit date: | 2000-05-23 | | Release date: | 2000-11-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nucleoside Binding Site of Herpes Simplex Type 1 Thymidine Kinase Analyzed by X-Ray Crystallography
Proteins: Struct.,Funct., Genet., 41, 2000
|
|
1E2I
 
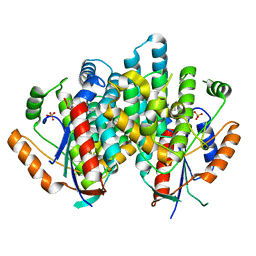 | | The nucleoside binding site of Herpes simplex type 1 thymidine kinase analyzed by X-ray crystallography | | Descriptor: | 9-HYDROXYPROPYLADENINE, R-ISOMER, S-ISOMER, ... | | Authors: | Vogt, J, Scapozza, L, Schulz, G.E. | | Deposit date: | 2000-05-23 | | Release date: | 2000-11-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nucleoside Binding Site of Herpes Simplex Type 1 Thymidine Kinase Analyzed by X-Ray Crystallography
Proteins, 41, 2000
|
|
6EZ8
 
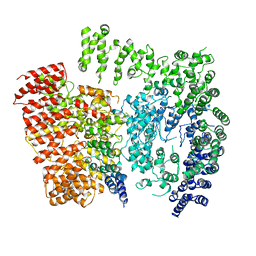 | | Human Huntingtin-HAP40 complex structure | | Descriptor: | Factor VIII intron 22 protein, Huntingtin | | Authors: | Guo, Q, Bin, H, Cheng, J, Pfeifer, G, Baumeister, W, Fernandez-Busnadiego, R, Kochanek, S. | | Deposit date: | 2017-11-14 | | Release date: | 2018-02-21 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The cryo-electron microscopy structure of huntingtin.
Nature, 555, 2018
|
|
7DXJ
 
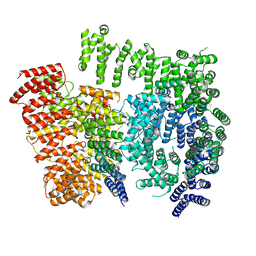 | |
7DXK
 
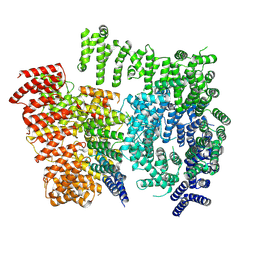 | |
3F0T
 
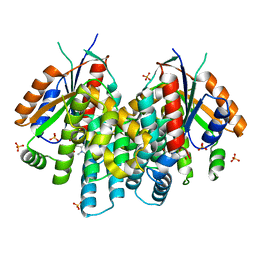 | | Crystal structure of thymidine kinase from Herpes simplex virus type 1 in complex with N-methyl-DHBT | | Descriptor: | N-Methyl-6-(1,3-dihydroxy-isobutyl)thymine, SULFATE ION, Thymidine kinase | | Authors: | Pernot, L, Perozzo, R, Westermaier, Y, Martic, M, Ametamey, S, Scapozza, L. | | Deposit date: | 2008-10-26 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis, crystal structure, and in vitro biological evaluation of C-6 pyrimidine derivatives: new lead structures for monitoring gene expression in vivo.
Nucleosides Nucleotides Nucleic Acids, 30, 2011
|
|
4R8T
 
 | | Structure of JEV protease | | Descriptor: | CHLORIDE ION, NS3, Serine protease subunit NS2B | | Authors: | Nair, D.T, Weinert, T, Wang, M, Olieric, V. | | Deposit date: | 2014-09-03 | | Release date: | 2014-12-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.133 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
4R8U
 
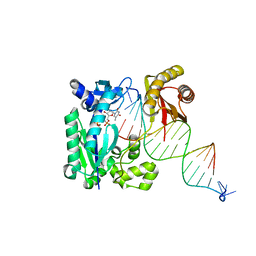 | | S-SAD structure of DINB-DNA Complex | | Descriptor: | 5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]thymidine, DNA, DNA polymerase IV, ... | | Authors: | Kottur, J, Nair, D.T, Weinert, T, Oligeric, V, Wang, M. | | Deposit date: | 2014-09-03 | | Release date: | 2015-01-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination
Nat.Methods, 12, 2015
|
|
4TNO
 
 | | Hypothetical protein PF1117 from Pyrococcus Furiosus: Structure solved by sulfur-SAD using Swiss Light Source Data | | Descriptor: | CHLORIDE ION, CRISPR-associated endoribonuclease Cas2 | | Authors: | Weinert, T, Waltersperger, S, Olieric, V, Panepucci, E, Chen, L, Rose, J.P, Wang, M, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2014-06-04 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
4PGO
 
 | | Crystal structure of hypothetical protein PF0907 from Pyrococcus furiosus solved by sulfur SAD using Swiss Light Source data | | Descriptor: | CHLORIDE ION, Uncharacterized protein | | Authors: | Weinert, T, Waltersperger, S, Olieric, V, Panepucci, E, Chen, L, Rose, J.P, Wang, M, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2014-05-02 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
4PII
 
 | | Crystal structure of hypothetical protein PF0907 from pyrococcus furiosus solved by sulfur SAD using Swiss light source data | | Descriptor: | CHLORIDE ION, IMIDAZOLE, N-glycosylase/DNA lyase | | Authors: | Weinert, T, Waltersperger, S, Olieric, V, Panepucci, E, Chen, L, Rose, J.P, Wang, M, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2014-05-08 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
4TN8
 
 | | Crystal structure of Thermus Thermophilus thioredoxin solved by sulfur SAD using Swiss Light Source data | | Descriptor: | CHLORIDE ION, Thioredoxin | | Authors: | Weinert, T, Waltersperger, S, Olieric, V, Panepucci, E, Chen, L, Rose, J.P, Wang, M, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2014-06-03 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
3RDP
 
 | | Crystal structure of thymidine kinase from herpes simplex virus type 1 in complex with N-METHYL-FHBT | | Descriptor: | 6-[(2R)-2-(fluoromethyl)-3-hydroxy-propyl]-1,5-dimethyl-pyrimidine-2,4-dione, SULFATE ION, Thymidine kinase | | Authors: | Pernot, L, Perozzo, R, Westermaier, Y, Martic, M, Ametamey, S, Scapozza, L. | | Deposit date: | 2011-04-01 | | Release date: | 2011-08-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Synthesis, crystal structure, and in vitro biological evaluation of C-6 pyrimidine derivatives: new lead structures for monitoring gene expression in vivo.
Nucleosides Nucleotides Nucleic Acids, 30, 2011
|
|
1QJ8
 
 | |
1QJ9
 
 | |
