6UFR
 
 | | Structure of recombinantly assembled E46K alpha-synuclein fibrils | | Descriptor: | Alpha-synuclein | | Authors: | Eisenberg, D.S, Boyer, D.R, Sawaya, M.R, Li, B, Jiang, L. | | Deposit date: | 2019-09-24 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | The alpha-synuclein hereditary mutation E46K unlocks a more stable, pathogenic fibril structure.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VW2
 
 | | Cryo-EM structure of human islet amyloid polypeptide (hIAPP, or amylin) fibrils | | Descriptor: | Islet amyloid polypeptide (SUMO-tagged) | | Authors: | Cao, Q, Boyer, D.R, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2020-02-18 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure and inhibitor design of human IAPP (amylin) fibrils.
Nat.Struct.Mol.Biol., 27, 2020
|
|
3ODV
 
 | | X-ray structure of kaliotoxin by racemic protein crystallography | | Descriptor: | CITRIC ACID, Potassium channel toxin alpha-KTx 3.1, trifluoroacetic acid | | Authors: | Pentelute, B.L, Mandal, K, Gates, Z.P, Sawaya, M.R, Yeates, T.O, Kent, S.B.H. | | Deposit date: | 2010-08-11 | | Release date: | 2010-10-20 | | Last modified: | 2012-03-28 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Total chemical synthesis and X-ray structure of kaliotoxin by racemic protein crystallography.
Chem.Commun.(Camb.), 46, 2010
|
|
3NVE
 
 | |
3SB7
 
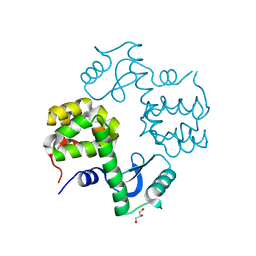 | | Cu-mediated Trimer of T4 Lysozyme D61H/K65H/R76H/R80H by Synthetic Symmetrization | | Descriptor: | COPPER (II) ION, GLYCEROL, Lysozyme | | Authors: | Soriaga, A.B, Laganowsky, A, Zhao, M, Sawaya, M.R, Cascio, D, Yeates, T.O. | | Deposit date: | 2011-06-03 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An approach to crystallizing proteins by metal-mediated synthetic symmetrization.
Protein Sci., 20, 2011
|
|
3OGI
 
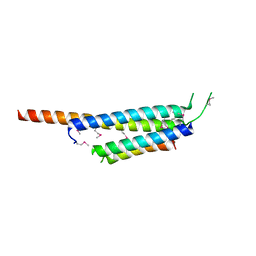 | | Crystal structure of the Mycobacterium tuberculosis H37Rv EsxOP complex (Rv2346c-Rv2347c) | | Descriptor: | Putative ESAT-6-like protein 6, Putative ESAT-6-like protein 7 | | Authors: | Arbing, M.A, Chan, S, Zhou, T.T, Ahn, C, Harris, L, Kuo, E, Sawaya, M.R, Cascio, D, Eisenberg, D, Integrated Center for Structure and Function Innovation (ISFI), TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-08-16 | | Release date: | 2010-08-25 | | Last modified: | 2014-05-21 | | Method: | X-RAY DIFFRACTION (2.549 Å) | | Cite: | Heterologous expression of mycobacterial Esx complexes in Escherichia coli for structural studies is facilitated by the use of maltose binding protein fusions.
Plos One, 8, 2013
|
|
3SB5
 
 | | Zn-mediated Trimer of T4 Lysozyme R125C/E128C by Synthetic Symmetrization | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme, ... | | Authors: | Laganowsky, A, Soriaga, A.B, Zhao, M, Sawaya, M.R, Cascio, D, Yeates, T.O. | | Deposit date: | 2011-06-03 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | An approach to crystallizing proteins by metal-mediated synthetic symmetrization.
Protein Sci., 20, 2011
|
|
3SGS
 
 | |
3SEX
 
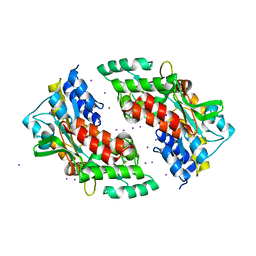 | | Ni-mediated Dimer of Maltose-binding Protein A216H/K220H by Synthetic Symmetrization (Form II) | | Descriptor: | IODIDE ION, Maltose-binding periplasmic protein, NICKEL (II) ION, ... | | Authors: | Zhao, M, Soriaga, A.B, Laganowsky, A, Sawaya, M.R, Cascio, D, Yeates, T.O. | | Deposit date: | 2011-06-11 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | An approach to crystallizing proteins by metal-mediated synthetic symmetrization.
Protein Sci., 20, 2011
|
|
3Q25
 
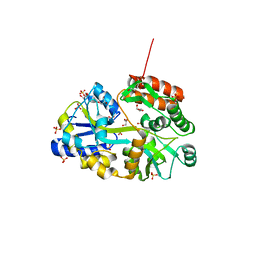 | | Crystal structure of human alpha-synuclein (1-19) fused to maltose binding protein (MBP) | | Descriptor: | GLYCEROL, Maltose-binding periplasmic protein/alpha-synuclein chimeric protein, SULFATE ION, ... | | Authors: | Zhao, M, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2010-12-19 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of segments of alpha-synuclein fused to maltose-binding protein suggest intermediate states during amyloid formation
Protein Sci., 20, 2011
|
|
3SB9
 
 | | Cu-mediated Dimer of T4 Lysozyme R76H/R80H by Synthetic Symmetrization | | Descriptor: | COPPER (II) ION, FORMIC ACID, Lysozyme | | Authors: | Soriaga, A.B, Laganowsky, A, Zhao, M, Sawaya, M.R, Cascio, D, Yeates, T.O. | | Deposit date: | 2011-06-03 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | An approach to crystallizing proteins by metal-mediated synthetic symmetrization.
Protein Sci., 20, 2011
|
|
3SEV
 
 | | Zn-mediated Trimer of Maltose-binding Protein E310H/K314H by Synthetic Symmetrization | | Descriptor: | CHLORIDE ION, Maltose-binding periplasmic protein, ZINC ION, ... | | Authors: | Zhao, M, Soriaga, A.B, Laganowsky, A, Sawaya, M.R, Cascio, D, Yeates, T.O. | | Deposit date: | 2011-06-11 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | An approach to crystallizing proteins by metal-mediated synthetic symmetrization.
Protein Sci., 20, 2011
|
|
3SB8
 
 | | Cu-mediated Dimer of T4 Lysozyme D61H/K65H by Synthetic Symmetrization | | Descriptor: | COPPER (II) ION, Lysozyme | | Authors: | Soriaga, A.B, Laganowsky, A, Zhao, M, Sawaya, M.R, Cascio, D, Yeates, T.O. | | Deposit date: | 2011-06-03 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | An approach to crystallizing proteins by metal-mediated synthetic symmetrization.
Protein Sci., 20, 2011
|
|
3Q9G
 
 | | VQIVY segment from Alzheimer's tau displayed on 42-membered macrocycle scaffold | | Descriptor: | ACETIC ACID, Cyclic pseudo-peptide VQIV(4BF)(ORN)(HAO)KL(ORN), GLYCEROL | | Authors: | Liu, C, Sawaya, M.R, Eisenberg, D, Nowick, J.S, Cheng, P, Zheng, J. | | Deposit date: | 2011-01-07 | | Release date: | 2011-06-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Characteristics of Amyloid-Related Oligomers Revealed by Crystal Structures of Macrocyclic beta-Sheet Mimics.
J.Am.Chem.Soc., 133, 2011
|
|
3Q9J
 
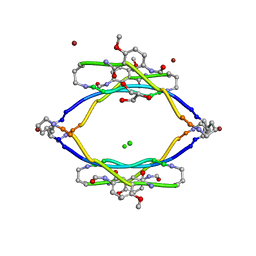 | | AIIFL segment derived from Alzheimer's Amyloid-Beta displayed on 42-membered macrocycle scaffold | | Descriptor: | CHLORIDE ION, Cyclic pseudo-peptide AIIFL(ORN)(HAO)YK(ORN), GLYCEROL, ... | | Authors: | Liu, C, Sawaya, M.R, Eisenberg, D, Nowick, J.S, Cheng, P, Zheng, J. | | Deposit date: | 2011-01-07 | | Release date: | 2011-06-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Characteristics of Amyloid-Related Oligomers Revealed by Crystal Structures of Macrocyclic beta-Sheet Mimics.
J.Am.Chem.Soc., 133, 2011
|
|
3SEY
 
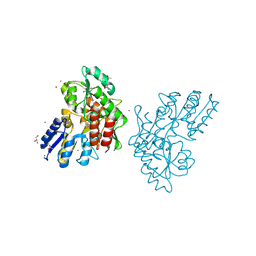 | | Zn-mediated Polymer of Maltose-binding Protein A216H/K220H by Synthetic Symmetrization (Form II) | | Descriptor: | ACETATE ION, GLYCEROL, Maltose-binding periplasmic protein, ... | | Authors: | Zhao, M, Soriaga, A.B, Laganowsky, A, Sawaya, M.R, Cascio, D, Yeates, T.O. | | Deposit date: | 2011-06-11 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An approach to crystallizing proteins by metal-mediated synthetic symmetrization.
Protein Sci., 20, 2011
|
|
3PZZ
 
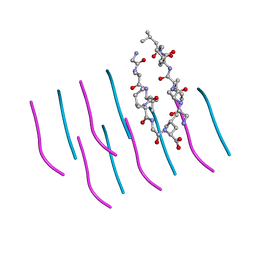 | |
3SBA
 
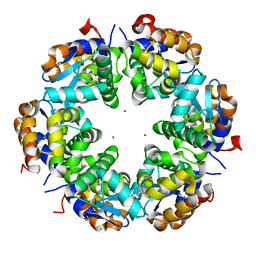 | | Zn-mediated Hexamer of T4 Lysozyme R76H/R80H by Synthetic Symmetrization | | Descriptor: | CHLORIDE ION, Lysozyme, ZINC ION | | Authors: | Soriaga, A.B, Laganowsky, A, Zhao, M, Sawaya, M.R, Cascio, D, Yeates, T.O. | | Deposit date: | 2011-06-03 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | An approach to crystallizing proteins by metal-mediated synthetic symmetrization.
Protein Sci., 20, 2011
|
|
3SES
 
 | | Cu-mediated Dimer of Maltose-binding Protein A216H/K220H by Synthetic Symmetrization | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Maltose-binding periplasmic protein, ... | | Authors: | Zhao, M, Soriaga, A.B, Laganowsky, A, Sawaya, M.R, Cascio, D, Yeates, T.O. | | Deposit date: | 2011-06-11 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An approach to crystallizing proteins by metal-mediated synthetic symmetrization.
Protein Sci., 20, 2011
|
|
3SET
 
 | | Ni-mediated Dimer of Maltose-binding Protein A216H/K220H by Synthetic Symmetrization (Form I) | | Descriptor: | Maltose-binding periplasmic protein, NICKEL (II) ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Zhao, M, Soriaga, A.B, Laganowsky, A, Sawaya, M.R, Cascio, D, Yeates, T.O. | | Deposit date: | 2011-06-11 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An approach to crystallizing proteins by metal-mediated synthetic symmetrization.
Protein Sci., 20, 2011
|
|
3SGO
 
 | |
3SKV
 
 | | Salicylyl-Acyltransferase SsfX3 from a Tetracycline Biosynthetic Pathway | | Descriptor: | GLYCEROL, SsfX3 | | Authors: | Pickens, L.B, Sawaya, M.R, Yeates, T.O, Tang, Y. | | Deposit date: | 2011-06-23 | | Release date: | 2011-10-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural and Biochemical Characterization of the Salicylyl-acyltranferase SsfX3 from a Tetracycline Biosynthetic Pathway.
J.Biol.Chem., 286, 2011
|
|
3SER
 
 | | Zn-mediated Polymer of Maltose-binding Protein K26H/K30H by Synthetic Symmetrization | | Descriptor: | CALCIUM ION, CHLORIDE ION, Maltose-binding periplasmic protein, ... | | Authors: | Zhao, M, Soriaga, A.B, Laganowsky, A, Sawaya, M.R, Cascio, D, Yeates, T.O. | | Deposit date: | 2011-06-11 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | An approach to crystallizing proteins by metal-mediated synthetic symmetrization.
Protein Sci., 20, 2011
|
|
3SGN
 
 | |
3NVF
 
 | |
