6KAF
 
 | | C2S2M2N2-type PSII-LHCII | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Chang, S.H, Shen, L.L, Huang, Z.H, Wang, W.D, Han, G.Y, Shen, J.R, Zhang, X. | | Deposit date: | 2019-06-22 | | Release date: | 2019-10-23 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Structure of a C2S2M2N2-type PSII-LHCII supercomplex from the green algaChlamydomonas reinhardtii.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7FIL
 
 | |
6Z3A
 
 | | Mec1-Ddc2 (wild-type) in complex with AMP-PNP | | Descriptor: | DNA damage checkpoint protein LCD1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase MEC1, ... | | Authors: | Yates, L.A, Zhang, X. | | Deposit date: | 2020-05-19 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Mechanism of auto-inhibition and activation of Mec1 ATR checkpoint kinase.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7WL2
 
 | |
7WLE
 
 | |
7WL9
 
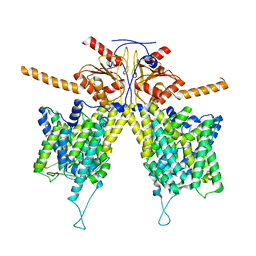 | | Mouse Pendrin in chloride and bicarbonate in asymmetric state | | Descriptor: | CHLORIDE ION, Pendrin | | Authors: | Liu, Q.Y, Zhang, X, Sun, L, Chen, Z.G. | | Deposit date: | 2022-01-12 | | Release date: | 2023-04-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Asymmetric pendrin homodimer reveals its molecular mechanism as anion exchanger.
Nat Commun, 14, 2023
|
|
7WLA
 
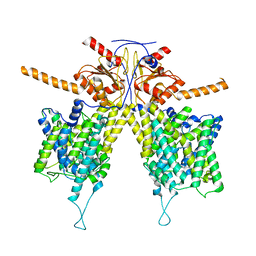 | |
7WK7
 
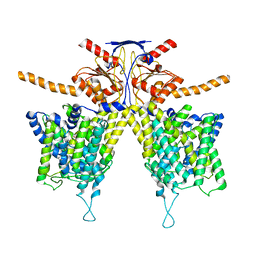 | | Mouse Pendrin bound bicarbonate in inward state | | Descriptor: | BICARBONATE ION, Pendrin | | Authors: | Liu, Q.Y, Zhang, X, Sun, L, Chen, Z.G. | | Deposit date: | 2022-01-08 | | Release date: | 2023-05-17 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Asymmetric pendrin homodimer reveals its molecular mechanism as anion exchanger.
Nat Commun, 14, 2023
|
|
7WK1
 
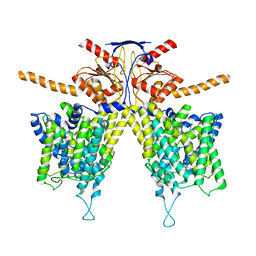 | | Mouse Pendrin bound chloride in inward state | | Descriptor: | CHLORIDE ION, Pendrin | | Authors: | Liu, Q.Y, Zhang, X, Sun, L, Chen, Z.G. | | Deposit date: | 2022-01-08 | | Release date: | 2023-04-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Asymmetric pendrin homodimer reveals its molecular mechanism as anion exchanger.
Nat Commun, 14, 2023
|
|
7WL7
 
 | |
7WL8
 
 | |
7WLB
 
 | |
7FBI
 
 | | Cryo-EM structure of EBV gB in complex with nAbs 3A3 and 3A5 | | Descriptor: | 3A3 heavy chain, 3A3 light chain, 3A5 heavy chain, ... | | Authors: | Zheng, Q, Li, S, Zha, Z, Hong, J, Chen, Y, Zhang, X. | | Deposit date: | 2021-07-10 | | Release date: | 2022-07-13 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of EBV gB in complex with nAbs 3A3 and 3A5
To Be Published
|
|
8BNZ
 
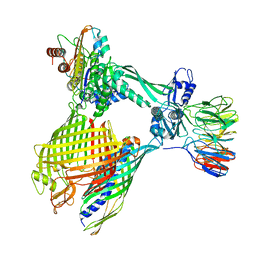 | | BAM-EspP complex structure with BamA-G431C/EspP-N1293C mutations in nanodisc | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Shen, C, Chang, S, Luo, Q, Zhang, Z, Xie, T, Luo, B, Lu, G, Zhu, X, Wei, X, Dong, C, Zhou, R, Zhang, X, Tang, X, Dong, H. | | Deposit date: | 2022-11-14 | | Release date: | 2023-04-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of BAM-mediated outer membrane beta-barrel protein assembly.
Nature, 617, 2023
|
|
8BO2
 
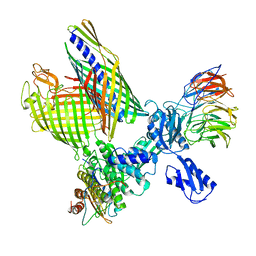 | | BAM-EspP complex structure with BamA-S425C/EspP-S1299C mutations in nanodisc | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Shen, C, Chang, S, Luo, Q, Zhang, Z, Xie, T, Luo, B, Lu, G, Zhu, X, Wei, X, Dong, C, Zhou, R, Zhang, X, Tang, X, Dong, H. | | Deposit date: | 2022-11-14 | | Release date: | 2023-04-26 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of BAM-mediated outer membrane beta-barrel protein assembly.
Nature, 617, 2023
|
|
8XGL
 
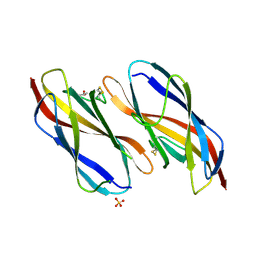 | | Structure of the Magnaporthe oryzae effector NIS1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Necrosis-inducing secreted protein 1, ... | | Authors: | Han, R, Wang, D, Zhang, X, Liu, J. | | Deposit date: | 2023-12-15 | | Release date: | 2024-09-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Understanding and manipulating the recognition of necrosis-inducing secreted protein 1 (NIS1) by BRI1-associated receptor kinase 1 (BAK1).
Int.J.Biol.Macromol., 278, 2024
|
|
6S8N
 
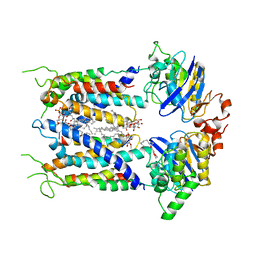 | | Cryo-EM structure of LptB2FGC in complex with lipopolysaccharide | | Descriptor: | 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, DODECYL-BETA-D-MALTOSIDE, Inner membrane protein yjgQ, ... | | Authors: | Tang, X.D, Chang, S.H, Luo, Q.H, Zhang, Z.Y, Qiao, W, Xu, C.H, Zhang, C.B, Niu, Y, Yang, W.X, Wang, T, Zhang, Z.B, Zhu, X.F, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2019-07-10 | | Release date: | 2019-09-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of lipopolysaccharide transporter LptB2FGC in lipopolysaccharide or AMP-PNP-bound states reveal its transport mechanism.
Nat Commun, 10, 2019
|
|
6S8H
 
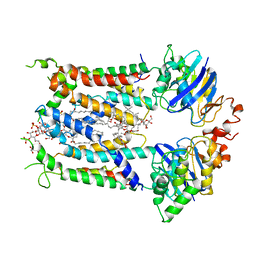 | | Cryo-EM structure of LptB2FG in complex with LPS | | Descriptor: | (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-[(2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-5-[(2~{S},3~{S},4~{R},5~{R},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-4-[(2~{R},3~{S},4~{R},5~{S},6~{R})-6-[(1~{S})-2-[(2~{S},3~{S},4~{S},5~{S},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-1-oxidanyl-ethyl]-3,4-bis(oxidanyl)-5-phosphonooxy-oxan-2-yl]oxy-3-oxidanyl-5-phosphonooxy-oxan-2-yl]oxy-2-carboxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[(3~{R})-3-dodecanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-3-oxidanyl-5-[[(3~{R})-3-oxidanyltetradecanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-4-[(3~{R})-3-tetradecanoyloxytetradecanoyl]oxy-oxan-2-yl]methoxy]oxan-4-yl]oxy-4,5-bis(oxidanyl)oxane-2-carboxylic acid, 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Tang, X.D, Chang, S.H, Luo, Q.H, Zhang, Z.Y, Qiao, W, Xu, C.H, Zhang, C.B, Niu, Y, Yang, W.X, Wang, T, Zhang, Z.B, Zhu, X.F, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2019-07-10 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of lipopolysaccharide transporter LptB2FGC in lipopolysaccharide or AMP-PNP-bound states reveal its transport mechanism.
Nat Commun, 10, 2019
|
|
6S8G
 
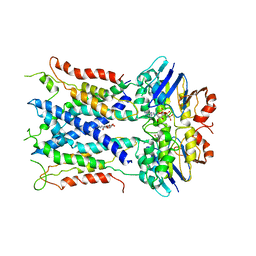 | | Cryo-EM structure of LptB2FGC in complex with AMP-PNP | | Descriptor: | Inner membrane protein yjgQ, LPS export ABC transporter permease LptF, Lipopolysaccharide ABC transporter, ... | | Authors: | Tang, X.D, Chang, S.H, Luo, Q.H, Zhang, Z.Y, Qiao, W, Xu, C.H, Zhang, C.B, Niu, Y, Yang, W.X, Wang, T, Zhang, Z.B, Zhu, X.F, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2019-07-10 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of lipopolysaccharide transporter LptB2FGC in lipopolysaccharide or AMP-PNP-bound states reveal its transport mechanism.
Nat Commun, 10, 2019
|
|
7BW7
 
 | | Cryo-EM Structure for the Ectodomain of the Full-length Human Insulin Receptor in Complex with 1 Insulin. | | Descriptor: | Insulin fusion, Insulin receptor | | Authors: | Yu, D, Zhang, X, Sun, J, Li, X, Wu, Z, Han, X, Fan, C, Ma, Y, Ouyang, Q, Wang, T. | | Deposit date: | 2020-04-13 | | Release date: | 2021-04-14 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Insulin Binding Induced the Ectodomain Conformational Dynamics in the Full-length Human Insulin Receptor
To Be Published
|
|
7BW8
 
 | | Cryo-EM Structure for the Insulin Binding Region in the Ectodomain of the Full-length Human Insulin Receptor in Complex with 1 Insulin | | Descriptor: | Insulin fusion, Insulin receptor | | Authors: | Yu, D, Zhang, X, Sun, J, Li, X, Wu, Z, Han, X, Fan, C, Ma, Y, Ouyang, Q, Wang, T. | | Deposit date: | 2020-04-14 | | Release date: | 2021-04-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Insulin Binding Induced the Ectodomain Conformational Dynamics in the Full-length Human Insulin Receptor
To Be Published
|
|
7BWA
 
 | | Cryo-EM Structure for the Ectodomain of the Full-length Human Insulin Receptor in Complex with 2 Insulin | | Descriptor: | Insulin fusion, Insulin receptor | | Authors: | Yu, D, Zhang, X, Sun, J, Li, X, Wu, Z, Han, X, Fan, C, Ma, Y, Ouyang, Q, Wang, T. | | Deposit date: | 2020-04-14 | | Release date: | 2021-04-14 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Insulin Binding Induced the Ectodomain Conformational Dynamics in the Full-length Human Insulin Receptor
To Be Published
|
|
7CCR
 
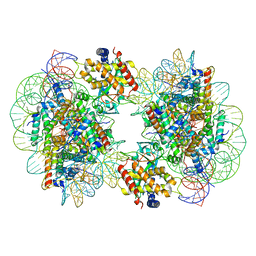 | | Structure of the 2:2 cGAS-nucleosome complex | | Descriptor: | Cyclic GMP-AMP synthase, DNA (147-MER), Histone H2A type 1-B/E, ... | | Authors: | Cao, D, Han, X, Fan, X, Xu, R.M, Zhang, X. | | Deposit date: | 2020-06-17 | | Release date: | 2020-10-07 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structural basis for nucleosome-mediated inhibition of cGAS activity.
Cell Res., 30, 2020
|
|
7CCQ
 
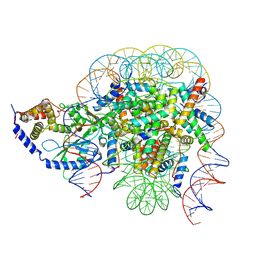 | | Structure of the 1:1 cGAS-nucleosome complex | | Descriptor: | Cyclic GMP-AMP synthase, DNA (147-MER), Histone H2A type 1-B/E, ... | | Authors: | Cao, D, Han, X, Fan, X, Xu, R.M, Zhang, X. | | Deposit date: | 2020-06-17 | | Release date: | 2020-10-07 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for nucleosome-mediated inhibition of cGAS activity.
Cell Res., 30, 2020
|
|
9JE9
 
 | | Crystal structure of a amidase that can hydrolase PU plastic | | Descriptor: | Amidase | | Authors: | Li, Z.S, Fang, S.T, Zheng, Z.R, Xia, W, Zhou, H.Y, Li, W.S, You, S, Han, X, Liu, W.D. | | Deposit date: | 2024-09-02 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Discovery and Engineering of a Urethanase for Enhanced Depolymerization of Polyurethane
Acs Catalysis, 15, 2025
|
|
