6FK7
 
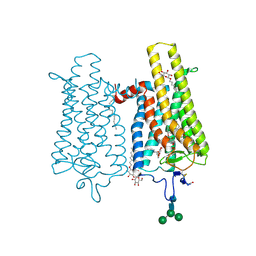 | | Crystal structure of N2C/D282C stabilized opsin bound to RS06 | | Descriptor: | (2~{R},3~{R})-2-(4-chlorophenyl)-3-oxidanyl-1-spiro[1,3-benzodioxole-2,4'-piperidine]-1'-yl-butan-1-one, PALMITIC ACID, Rhodopsin, ... | | Authors: | Mattle, D, Standfuss, J, Dawson, R. | | Deposit date: | 2018-01-23 | | Release date: | 2018-04-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Ligand channel in pharmacologically stabilized rhodopsin.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7MHG
 
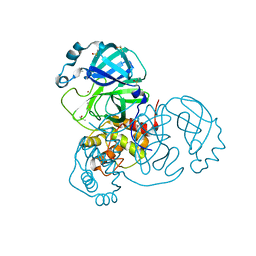 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) at 240 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5302 Å) | | Cite: | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7MHL
 
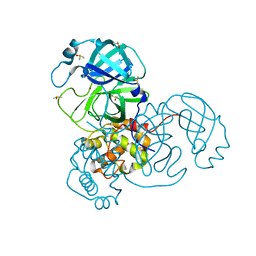 | | Ensemble refinement structure of SARS-CoV-2 main protease (Mpro) at 100 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7MHM
 
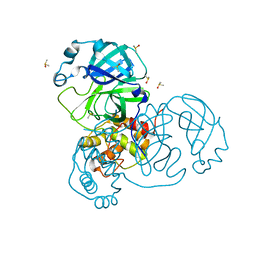 | | Ensemble refinement structure of SARS-CoV-2 main protease (Mpro) at 240 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5302 Å) | | Cite: | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
6G5T
 
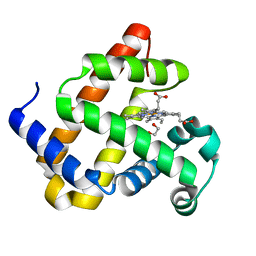 | | Myoglobin H64V/V68A in the resting state, 1.5 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tinzl, M, Hayashi, T, Mori, T, Hilvert, D. | | Deposit date: | 2018-03-30 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Capture and characterization of a reactive haem-carbenoid complex in an artificial metalloenzyme
Nat Catal, 1, 2018
|
|
6T8U
 
 | | Complement factor B in complex with 5-Bromo-3-chloro-N-(4,5-dihydro-1H-imidazol-2-yl)-7-methyl-1H-indol-4-amine | | Descriptor: | 5-bromanyl-3-chloranyl-~{N}-(1~{H}-imidazol-2-yl)-7-methyl-1~{H}-indol-4-amine, Complement factor B, SULFATE ION | | Authors: | Mainolfi, N, Ehara, T, Karki, R.G, Anderson, K, Mac Sweeney, A, Wiesmann, C, Adams, C, Liao, S.-M, Argikar, U.A, Jendza, K, Zhang, C, Powers, J, Klosowski, D.W, Crowley, M, Kawanami, T, Ding, J, April, M, Forster, C, Serrano-Wu, M, Capparelli, M, Ramqaj, R, Solovay, C, Cumin, F, Smith, T.M, Ferrara, L, Lee, W, Long, D, Prentiss, M, De Erkenez, A, Yang, L, Fang, L, Sellner, H, Sirockin, F, Valeur, E, Erbel, P, Ramage, P, Gerhartz, B, Schubart, A, Flohr, S, Gradoux, N, Feifel, R, Vogg, B, Maibaum, J, Eder, J, Sedrani, R, Harrison, R.A, Mogi, M, Jaffee, B.D, Adams, C.M. | | Deposit date: | 2019-10-25 | | Release date: | 2020-03-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Discovery of 4-((2S,4S)-4-Ethoxy-1-((5-methoxy-7-methyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic Acid (LNP023), a Factor B Inhibitor Specifically Designed To Be Applicable to Treating a Diverse Array of Complement Mediated Diseases.
J.Med.Chem., 63, 2020
|
|
6DLC
 
 | |
3HLJ
 
 | | Crystal structure of human carbonic anhydrase isozyme II with 3-methylthiobenzimidazo[1,2-c][1,2,3]thiadiazol-7-sulfonamide | | Descriptor: | 3-methylthiobenzimidazo[1,2-c][1,2,3]thiadiazol-7-sulfonamide, Carbonic anhydrase 2, SODIUM ION, ... | | Authors: | Grazulis, S, Manakova, E, Golovenko, D. | | Deposit date: | 2009-05-27 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Inhibition and binding studies of carbonic anhydrase isozymes I, II and IX with benzimidazo[1,2-c][1,2,3]thiadiazole-7-sulphonamides
J Enzyme Inhib Med Chem, 25, 2010
|
|
2XSL
 
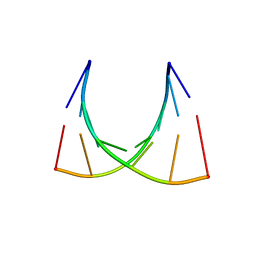 | | The crystal structure of a Thermus thermophilus tRNAGly acceptor stem microhelix at 1.6 Angstroem resolution | | Descriptor: | 5'-R(*CP*UP*CP*CP*CP*GP*C)-3', 5'-R(*GP*CP*GP*GP*GP*AP*G)-3' | | Authors: | Oberthuer, D, Eichert, A, Erdmann, V.A, Fuerste, J.P, Betzel, C, Foerster, C. | | Deposit date: | 2010-10-29 | | Release date: | 2011-08-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | The Crystal Structure of a Thermus Thermophilus tRNA(Gly) Acceptor Stem Microhelix at 1.6 A Resolution.
Biochem.Biophys.Res.Commun., 404, 2011
|
|
4AK9
 
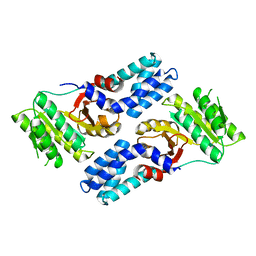 | |
3ZUJ
 
 | | Padron on (fluorescent) ABcis | | Descriptor: | FLUORESCENT PROTEIN DRONPA | | Authors: | REGIS Faro, A, Carpentier, P, Bourgeois, D. | | Deposit date: | 2011-07-19 | | Release date: | 2011-08-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.345 Å) | | Cite: | Low-Temperature Chromophore Isomerization Reveals the Photoswitching Mechanism of the Fluorescent Protein Padron.
J.Am.Chem.Soc., 133, 2011
|
|
2Y5C
 
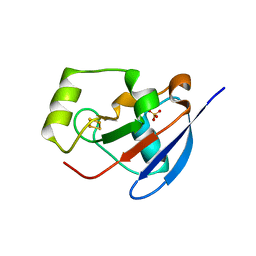 | | Structure of human ferredoxin 2 (Fdx2)in complex with 2Fe2S cluster | | Descriptor: | ADRENODOXIN-LIKE PROTEIN, MITOCHONDRIAL, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Webert, H, Hobler, A, Sheftel, A.D, Molik, S, Maestre-Reyna, M, Essen, L.-O, Vorniscescu, D, Keusgen, M, Hannemann, F, Bernhardt, R, Lill, R. | | Deposit date: | 2011-01-12 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and Functional Studies on Human Mitochondrial Ferredoxins
To be Published
|
|
6YJM
 
 | | Crystal Structure of the Catalytic Domain of ADAMTS-5 in Complex with the Inhibitor GLPG1972 | | Descriptor: | (5~{S})-5-[3-[(3~{S})-4-[3,5-bis(fluoranyl)phenyl]-3-methyl-piperazin-1-yl]-3-oxidanylidene-propyl]-5-cyclopropyl-imidazolidine-2,4-dione, A disintegrin and metalloproteinase with thrombospondin motifs 5, CALCIUM ION, ... | | Authors: | Goepfert, A, Leonard, P, Triballeau, N, Fleury, D, Mollat, P, Lamers, M. | | Deposit date: | 2020-04-03 | | Release date: | 2021-04-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Discovery of GLPG1972/S201086, a Potent, Selective, and Orally Bioavailable ADAMTS-5 Inhibitor for the Treatment of Osteoarthritis.
J.Med.Chem., 64, 2021
|
|
6TGX
 
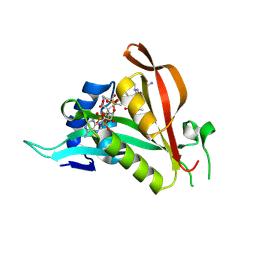 | | Crystal structure of Arabidopsis thaliana NAA60 in complex with a bisubstrate analogue | | Descriptor: | Acyl-CoA N-acyltransferases (NAT) superfamily protein, CARBOXYMETHYL COENZYME *A, MET-VAL-ASN-ALA | | Authors: | Layer, D, Kopp, J, Lapouge, K, Sinning, I. | | Deposit date: | 2019-11-18 | | Release date: | 2020-06-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The Arabidopsis N alpha -acetyltransferase NAA60 locates to the plasma membrane and is vital for the high salt stress response.
New Phytol., 228, 2020
|
|
2OSE
 
 | | Crystal Structure of the Mimivirus Cyclophilin | | Descriptor: | CHLORIDE ION, Probable peptidyl-prolyl cis-trans isomerase | | Authors: | Eisenmesser, E.Z, Thai, V, Renesto, P, Raoult, D. | | Deposit date: | 2007-02-05 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural, biochemical, and in vivo characterization of the first virally encoded cyclophilin from the Mimivirus.
J.Mol.Biol., 378, 2008
|
|
1LIT
 
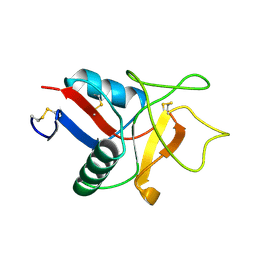 | | HUMAN LITHOSTATHINE | | Descriptor: | LITHOSTATHINE | | Authors: | Bertrand, J.A, Pignol, D, Bernard, J.-P, Verdier, J.-M, Dagorn, J.-C, Fontacilla-Camps, J.C. | | Deposit date: | 1996-01-17 | | Release date: | 1997-01-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of human lithostathine, the pancreatic inhibitor of stone formation.
EMBO J., 15, 1996
|
|
1GT8
 
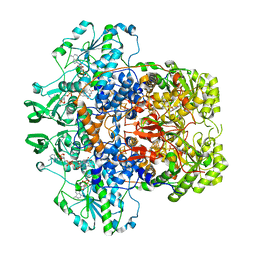 | | DIHYDROPYRIMIDINE DEHYDROGENASE (DPD) FROM PIG, TERNARY COMPLEX WITH NADPH AND URACIL-4-ACETIC ACID | | Descriptor: | DIHYDROPYRIMIDINE DEHYDROGENASE, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Dobritzsch, D, Ricagno, S, Schneider, G, Schnackerz, K.D, Lindqvist, Y. | | Deposit date: | 2002-01-14 | | Release date: | 2002-04-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the productive ternary complex of dihydropyrimidine dehydrogenase with NADPH and 5-iodouracil. Implications for mechanism of inhibition and electron transfer.
J. Biol. Chem., 277, 2002
|
|
3HVH
 
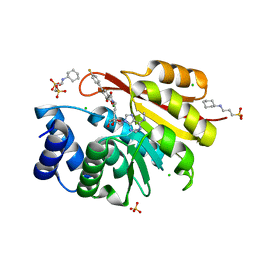 | | Rat catechol O-methyltransferase in complex with a catechol-type, N6-methyladenine-containing bisubstrate inhibitor | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, CHLORIDE ION, Catechol O-methyltransferase, ... | | Authors: | Ehler, A, Schlatter, D, Stihle, M, Benz, J, Rudolph, M.G. | | Deposit date: | 2009-06-16 | | Release date: | 2009-10-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular recognition at the active site of catechol-o-methyltransferase: energetically favorable replacement of a water molecule imported by a bisubstrate inhibitor.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
1GTH
 
 | | DIHYDROPYRIMIDINE DEHYDROGENASE (DPD) FROM PIG, TERNARY COMPLEX WITH NADPH AND 5-IODOURACIL | | Descriptor: | (5S)-5-IODODIHYDRO-2,4(1H,3H)-PYRIMIDINEDIONE, 5-IODOURACIL, DIHYDROPYRIMIDINE DEHYDROGENASE, ... | | Authors: | Dobritzsch, D, Ricagno, S, Schneider, G, Schnackerz, K.D, Lindqvist, Y. | | Deposit date: | 2002-01-15 | | Release date: | 2002-04-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the productive ternary complex of dihydropyrimidine dehydrogenase with NADPH and 5-iodouracil. Implications for mechanism of inhibition and electron transfer.
J. Biol. Chem., 277, 2002
|
|
6N9F
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with a short substrate mimic ACCA-DPhe and bound to mRNA and P-site tRNA at 3.7A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S19, ... | | Authors: | Melnikov, S.V, Khabibullina, N.F, Mairhofer, E, Vargas-Rodriguez, O, Reynolds, N.M, Micura, R, Soll, D, Polikanov, Y.S. | | Deposit date: | 2018-12-03 | | Release date: | 2018-12-12 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Mechanistic insights into the slow peptide bond formation with D-amino acids in the ribosomal active site.
Nucleic Acids Res., 47, 2019
|
|
8FMX
 
 | | Langya virus F glycoprotein ectodomain in prefusion form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Low, Y.S, Isaacs, A, Modhiran, N, Watterson, D. | | Deposit date: | 2022-12-26 | | Release date: | 2023-06-28 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structure and antigenicity of divergent Henipavirus fusion glycoproteins.
Nat Commun, 14, 2023
|
|
1GTE
 
 | | DIHYDROPYRIMIDINE DEHYDROGENASE (DPD) FROM PIG, BINARY COMPLEX WITH 5-IODOURACIL | | Descriptor: | 5-IODOURACIL, DIHYDROPYRIMIDINE DEHYDROGENASE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Dobritzsch, D, Ricagno, S, Schneider, G, Schnackerz, K.D, Lindqvist, Y. | | Deposit date: | 2002-01-15 | | Release date: | 2002-04-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the productive ternary complex of dihydropyrimidine dehydrogenase with NADPH and 5-iodouracil. Implications for mechanism of inhibition and electron transfer.
J. Biol. Chem., 277, 2002
|
|
8FMY
 
 | |
5J6Q
 
 | | Cwp8 from Clostridium difficile | | Descriptor: | CHLORIDE ION, Cell wall binding protein cwp8, SULFATE ION | | Authors: | Renko, M, Usenik, A, Turk, D. | | Deposit date: | 2016-04-05 | | Release date: | 2017-02-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The CWB2 Cell Wall-Anchoring Module Is Revealed by the Crystal Structures of the Clostridium difficile Cell Wall Proteins Cwp8 and Cwp6.
Structure, 25, 2017
|
|
2P4L
 
 | | Structure and sodium channel activity of an excitatory I1-superfamily conotoxin | | Descriptor: | I-superfamily conotoxin r11a | | Authors: | Buczek, O, Wei, D.X, Babon, J.J, Yang, X.D, Fiedler, B, Yoshikami, D, Olivera, B.M, Bulaj, G, Norton, R.S. | | Deposit date: | 2007-03-12 | | Release date: | 2007-09-25 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structure and Sodium Channel Activity of an Excitatory I(1)-Superfamily Conotoxin
Biochemistry, 46, 2007
|
|
