8UVL
 
 | | Crystal structure of selective IRE1a inhibitor 29 at the enzyme active site | | Descriptor: | 1,2-ETHANEDIOL, 1-phenyl-N-(2,3,6-trifluoro-4-{[(3M)-3-(2-{[(3R,5R)-5-fluoropiperidin-3-yl]amino}pyrimidin-4-yl)pyridin-2-yl]oxy}phenyl)methanesulfonamide, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Kiefer, J.R, Wallweber, H.A, Braun, M.-G, Wei, W, Jiang, F, Wang, W, Rudolph, J, Ashkenazi, A. | | Deposit date: | 2023-11-03 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Discovery of Potent, Selective, and Orally Available IRE1 alpha Inhibitors Demonstrating Comparable PD Modulation to IRE1 Knockdown in a Multiple Myeloma Model.
J.Med.Chem., 67, 2024
|
|
5WBS
 
 | | Crystal structure of Frizzled-7 CRD with an inhibitor peptide Fz7-21 | | Descriptor: | Frizzled-7,inhibitor peptide Fz7-21 | | Authors: | Nile, A.H, Mukund, S, Hannoush, R.N, Wang, W. | | Deposit date: | 2017-06-29 | | Release date: | 2018-04-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | A selective peptide inhibitor of Frizzled 7 receptors disrupts intestinal stem cells.
Nat. Chem. Biol., 14, 2018
|
|
5URV
 
 | | Crystal structure of Frizzled 7 CRD in complex with C24 fatty acid | | Descriptor: | (15E)-TETRACOS-15-ENOIC ACID, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mukund, S, Nile, A.H, Stanger, K, Hannoush, R.N, Wang, W. | | Deposit date: | 2017-02-13 | | Release date: | 2017-05-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unsaturated fatty acyl recognition by Frizzled receptors mediates dimerization upon Wnt ligand binding.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5URZ
 
 | | Crystal structure of Frizzled 5 CRD in complex with BOG | | Descriptor: | Frizzled-5, alpha-L-fucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, octyl beta-D-glucopyranoside | | Authors: | Mukund, S, Nile, A.H, Stanger, K, Hannoush, R.N, Wang, W. | | Deposit date: | 2017-02-13 | | Release date: | 2017-05-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Unsaturated fatty acyl recognition by Frizzled receptors mediates dimerization upon Wnt ligand binding.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5URY
 
 | | Crystal structure of Frizzled 5 CRD in complex with PAM | | Descriptor: | Frizzled-5, PALMITOLEIC ACID, alpha-L-fucopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)][alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Mukund, S, Nile, A.H, Stanger, K, Hannoush, R.N, Wang, W. | | Deposit date: | 2017-02-13 | | Release date: | 2017-05-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Unsaturated fatty acyl recognition by Frizzled receptors mediates dimerization upon Wnt ligand binding.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VZY
 
 | | Crystal structure of crenezumab Fab in complex with Abeta | | Descriptor: | Amyloid beta A4 protein, Crenezumab Fab heavy chain,Immunoglobulin gamma-1 heavy chain, Crenezumab Fab light chain,Immunoblobulin light chain | | Authors: | Ultsch, M, Wang, W. | | Deposit date: | 2017-05-29 | | Release date: | 2017-08-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure of Crenezumab Complex with Abeta Shows Loss of beta-Hairpin.
Sci Rep, 6, 2016
|
|
5VRQ
 
 | | Crystal structure of Legionella pneumophila effector AnkC | | Descriptor: | Ankyrin repeat-containing protein | | Authors: | Kozlov, G, Wong, K, Wang, W, Skubak, P, Munoz-Escobar, J, Liu, Y, Pannu, N.S, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2017-05-11 | | Release date: | 2017-11-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.205 Å) | | Cite: | Ankyrin repeats as a dimerization module.
Biochem. Biophys. Res. Commun., 495, 2018
|
|
5W96
 
 | | Solution structure of phage derived peptide inhibitor of frizzled 7 receptor | | Descriptor: | Fz7 binding peptide | | Authors: | Nile, A.H, de Sousa e Melo, F, Mukund, S, Piskol, R, Hansen, S, Zhou, L, Zhang, Y, Fu, Y, Gogol, E.B, Komuves, L.G, Modrusan, Z, Angers, S, Franke, Y, Koth, C, Fairbrother, W.J, Wang, W, de Sauvage, F.J, Hannoush, R.N. | | Deposit date: | 2017-06-22 | | Release date: | 2018-04-18 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | A selective peptide inhibitor of Frizzled 7 receptors disrupts intestinal stem cells.
Nat. Chem. Biol., 14, 2018
|
|
7K0V
 
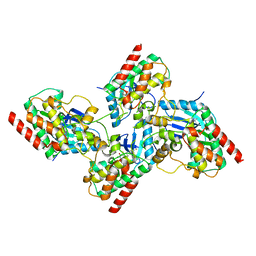 | | Crystal structure of bRaf in complex with inhibitor GNE-0749 | | Descriptor: | CHLORIDE ION, N-(3,3-dimethylbutyl)-N'-{2-fluoro-5-[(5-fluoro-3-methyl-4-oxo-3,4-dihydroquinazolin-6-yl)amino]-4-methylphenyl}urea, Non-specific serine/threonine protein kinase | | Authors: | Yin, J, Eigenbrot, C.E, Wang, W. | | Deposit date: | 2020-09-06 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Targeting KRAS Mutant Cancers via Combination Treatment: Discovery of a 5-Fluoro-4-(3 H )-quinazolinone Aryl Urea pan-RAF Kinase Inhibitor.
J.Med.Chem., 64, 2021
|
|
7CSZ
 
 | | Crystal structure of the N-terminal tandem RRM domains of RBM45 in complex with single-stranded DNA | | Descriptor: | DNA (5'-D(*CP*GP*AP*CP*GP*GP*GP*AP*CP*GP*C)-3'), RNA-binding protein 45 | | Authors: | Chen, X, Yang, Z, Wang, W, Wang, M. | | Deposit date: | 2020-08-17 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for RNA recognition by the N-terminal tandem RRM domains of human RBM45.
Nucleic Acids Res., 49, 2021
|
|
7CSX
 
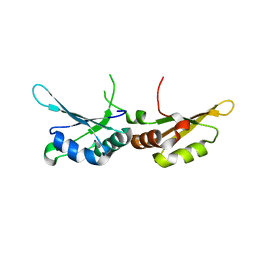 | |
7E2P
 
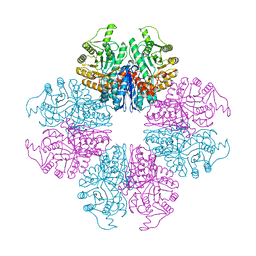 | | The Crystal Structure of Mycoplasma bovis enolase | | Descriptor: | Enolase | | Authors: | Chen, R, Zhang, S, Gan, R, Wang, W, Ran, T, Shao, G, Xiong, Q, Feng, Z. | | Deposit date: | 2021-02-07 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evidence for the Rapid and Divergent Evolution of Mycoplasmas: Structural and Phylogenetic Analysis of Enolases.
Front Mol Biosci, 8, 2022
|
|
7E2Q
 
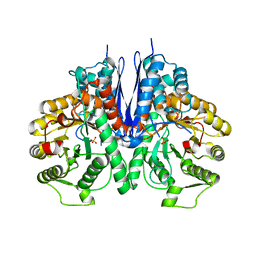 | | Crystal structure of Mycoplasma pneumoniae Enolase | | Descriptor: | Enolase, SULFATE ION | | Authors: | Chen, R, Zhang, S, Gan, R, Wang, W, Ran, T, Xiong, Q, Shao, G, Feng, Z. | | Deposit date: | 2021-02-07 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evidence for the Rapid and Divergent Evolution of Mycoplasmas: Structural and Phylogenetic Analysis of Enolases.
Front Mol Biosci, 8, 2022
|
|
7WN8
 
 | | Crystal structure of antibody (BC31M5) binds to CD47 | | Descriptor: | BC31M5 Fab Heavy chain, BC31M5 Fab Light chain, Leukocyte surface antigen CD47, ... | | Authors: | Li, Y, Wang, W, Sui, J, Zhang, S. | | Deposit date: | 2022-01-17 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A pH-dependent anti-CD47 antibody that selectively targets solid tumors and improves therapeutic efficacy and safety.
J Hematol Oncol, 16, 2023
|
|
8I79
 
 | | Cryo-EM structure of KCTD7 in complex with Cullin3 | | Descriptor: | BTB/POZ domain-containing protein KCTD7, Cullin-3 | | Authors: | Jiang, W, Wang, W, Zheng, S. | | Deposit date: | 2023-01-31 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for the ubiquitination of G protein beta gamma subunits by KCTD5/Cullin3 E3 ligase.
Sci Adv, 9, 2023
|
|
6JLU
 
 | | Structure of PSII-FCP supercomplex from a centric diatom Chaetoceros gracilis at 3.02 angstrom resolution | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'-yl acetate, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Pi, X, Zhao, S, Wang, W, Kuang, T, Sui, S, Shen, J. | | Deposit date: | 2019-03-06 | | Release date: | 2019-07-31 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | The pigment-protein network of a diatom photosystem II-light-harvesting antenna supercomplex.
Science, 365, 2019
|
|
7DKZ
 
 | | Structure of plant photosystem I-light harvesting complex I supercomplex | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Wang, J, Yu, L.J, Wang, W. | | Deposit date: | 2020-11-25 | | Release date: | 2021-03-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Structure of plant photosystem I-light harvesting complex I supercomplex at 2.4 angstrom resolution.
J Integr Plant Biol, 63, 2021
|
|
8J0D
 
 | | FCP heterodimer, Lhca2, and Lhcf5 together as the M1 side binds to the PSII core in the diatom Thalassiosira pseudonana | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Li, Z, Feng, Y, Wang, W, Shen, J.R. | | Deposit date: | 2023-04-10 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structure of a diatom photosystem II supercomplex containing a member of Lhcx family and dimeric FCPII.
Sci Adv, 9, 2023
|
|
7F79
 
 | | Crystal structure of glutamate dehydrogenase 3 from Candida albicans in complex with alpha-ketoglutarate and NADPH | | Descriptor: | 2-OXOGLUTARIC ACID, GLYCEROL, Glutamate dehydrogenase, ... | | Authors: | Li, N, Wang, W, Zeng, X, Liu, M, Li, M, Li, C, Wang, M. | | Deposit date: | 2021-06-28 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of glutamate dehydrogenase 3 from Candida albicans.
Biochem.Biophys.Res.Commun., 570, 2021
|
|
7F77
 
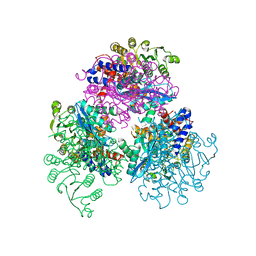 | | Crystal structure of glutamate dehydrogenase 3 from Candida albicans | | Descriptor: | Glutamate dehydrogenase | | Authors: | Li, N, Wang, W, Zeng, X, Liu, M, Li, M, Li, C, Wang, M. | | Deposit date: | 2021-06-28 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.086 Å) | | Cite: | Crystal structure of glutamate dehydrogenase 3 from Candida albicans.
Biochem.Biophys.Res.Commun., 570, 2021
|
|
8HEH
 
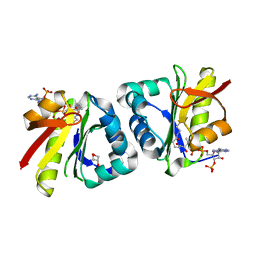 | | Crystal structure of GCN5-related N-acetyltransferase 05790 | | Descriptor: | COENZYME A, GLYCEROL, GNAT family N-acetyltransferase | | Authors: | Xu, M.X, Ran, T.T, Wang, W. | | Deposit date: | 2022-11-08 | | Release date: | 2022-12-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of prodigiosin binding protein PgbP, a GNAT family protein, in Serratia marcescens FS14.
Biochem.Biophys.Res.Commun., 640, 2022
|
|
8H0M
 
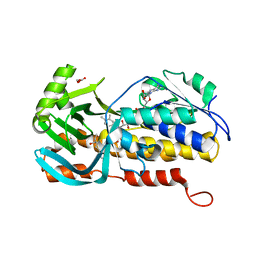 | | Crystal structure of VioD | | Descriptor: | (2S)-2-ethylhexan-1-ol, FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, ... | | Authors: | Xu, M, Ran, T, Wang, W. | | Deposit date: | 2022-09-29 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Structural basis for substrate binding and catalytic mechanism of the key enzyme VioD in the violacein synthesis pathway.
Proteins, 91, 2023
|
|
6XDF
 
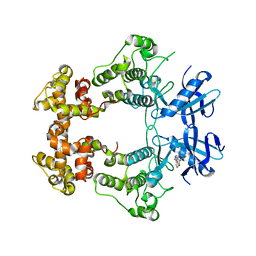 | | Crystal structure of IRE1a in complex with G-4100 | | Descriptor: | 4-amino-N-(6-chloro-2-fluoro-3-{[(pyrrolidin-1-yl)sulfonyl]amino}phenyl)quinazoline-8-carboxamide, SODIUM ION, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Steinbacher, S, Wang, W. | | Deposit date: | 2020-06-10 | | Release date: | 2021-04-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Identification of BRaf-Sparing Amino-Thienopyrimidines with Potent IRE1 alpha Inhibitory Activity.
Acs Med.Chem.Lett., 11, 2020
|
|
6XDD
 
 | | Crystal structure of IRE1 in complex with G-3053 | | Descriptor: | 4-[(trans-4-aminocyclohexyl)amino]-N-(6-chloro-3-{[(2,5-difluorophenyl)sulfonyl]amino}-2-fluorophenyl)thieno[3,2-d]pyrimidine-7-carboxamide, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Ackerly-Wallweber, H, Wang, W. | | Deposit date: | 2020-06-10 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of BRaf-Sparing Amino-Thienopyrimidines with Potent IRE1 alpha Inhibitory Activity.
Acs Med.Chem.Lett., 11, 2020
|
|
8H4E
 
 | | Blasnase-T13A/P55N with D-asn | | Descriptor: | D-ASPARAGINE, FORMIC ACID, L-asparaginase, ... | | Authors: | Lu, F, Wang, W, Chi, H, Ran, T. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structure-based rational design of Bacillus licheniformis L-asparaginase with low/no D-asparaginase activity for a safer enzyme
To Be Published
|
|
