7CCS
 
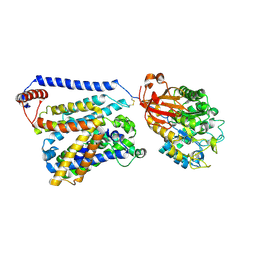 | | Consensus mutated xCT-CD98hc complex | | Descriptor: | 4F2 cell-surface antigen heavy chain, Consensus mutated Anionic Amino Acid Transporter Light Chain, Xc- System | | Authors: | Oda, K, Lee, Y, Takemoto, M, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2020-06-17 | | Release date: | 2020-12-09 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Consensus mutagenesis approach improves the thermal stability of system x c - transporter, xCT, and enables cryo-EM analyses.
Protein Sci., 29, 2020
|
|
2ZNI
 
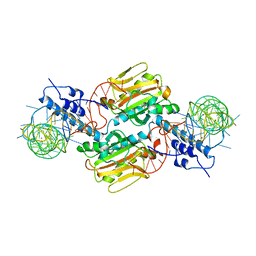 | | Crystal structure of Pyrrolysyl-tRNA synthetase-tRNA(Pyl) complex from Desulfitobacterium hafniense | | Descriptor: | CALCIUM ION, Pyrrolysyl-tRNA synthetase, bacterial tRNA | | Authors: | Nozawa, K, Araiso, Y, Soll, D, Ishitani, R, Nureki, O. | | Deposit date: | 2008-04-25 | | Release date: | 2008-12-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Pyrrolysyl-tRNA synthetase-tRNA(Pyl) structure reveals the molecular basis of orthogonality
Nature, 457, 2009
|
|
2ZNV
 
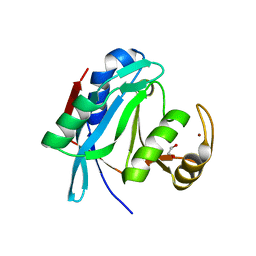 | | Crystal structure of human AMSH-LP DUB domain in complex with Lys63-linked ubiquitin dimer | | Descriptor: | 1,2-ETHANEDIOL, AMSH-like protease, Ubiquitin, ... | | Authors: | Sato, Y, Azusa, Y, Yamagata, A, Mimura, H, Wang, X, Yamashita, M, Ookata, K, Nureki, O, Iwai, K, Komada, M, Fukai, S. | | Deposit date: | 2008-05-01 | | Release date: | 2008-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains
Nature, 455, 2008
|
|
2ZW9
 
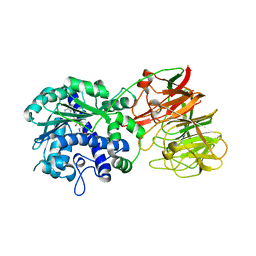 | | Crystal structure of tRNA wybutosine synthesizing enzyme TYW4 | | Descriptor: | Leucine carboxyl methyltransferase 2, S-ADENOSYLMETHIONINE | | Authors: | Suzuki, Y, Noma, A, Suzuki, T, Ishitani, R, Nureki, O. | | Deposit date: | 2008-12-01 | | Release date: | 2009-06-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of tRNA modification with CO2 fixation and methylation by wybutosine synthesizing enzyme TYW4.
Nucleic Acids Res., 37, 2009
|
|
2Z2U
 
 | | Crystal structure of archaeal TYW1 | | Descriptor: | UPF0026 protein MJ0257 | | Authors: | Suzuki, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2007-05-28 | | Release date: | 2007-10-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Radical SAM Enzyme Catalyzing Tricyclic Modified Base Formation in tRNA
J.Mol.Biol., 372, 2007
|
|
2ZNR
 
 | | Crystal structure of the DUB domain of human AMSH-LP | | Descriptor: | 1,2-ETHANEDIOL, AMSH-like protease, PRASEODYMIUM ION, ... | | Authors: | Sato, Y, Azusa, Y, Yamagata, A, Mimura, H, Wang, X, Yamashita, M, Ookata, K, Nureki, O, Iwai, K, Komada, M, Fukai, S. | | Deposit date: | 2008-05-01 | | Release date: | 2008-09-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains
Nature, 455, 2008
|
|
3A1U
 
 | | Crystal structue of the cytosolic domain of T. maritima FeoB iron iransporter in GMPPNP form | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Iron(II) transport protein B, MAGNESIUM ION, ... | | Authors: | Hattori, M, Ishitani, R, Nureki, O. | | Deposit date: | 2009-04-22 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of novel interactions between the small-GTPase and GDI-like domains in prokaryotic FeoB iron transporter
Structure, 17, 2009
|
|
3A27
 
 | | Crystal structure of M. jannaschii TYW2 in complex with AdoMet | | Descriptor: | S-ADENOSYLMETHIONINE, Uncharacterized protein MJ1557 | | Authors: | Umitsu, M, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2009-04-28 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Structural basis of AdoMet-dependent aminocarboxypropyl transfer reaction catalyzed by tRNA-wybutosine synthesizing enzyme, TYW2
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3A2K
 
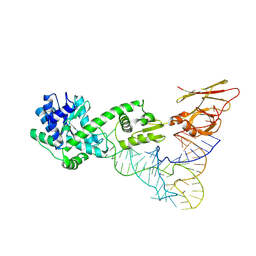 | | Crystal structure of TilS complexed with tRNA | | Descriptor: | bacterial tRNA, tRNA(Ile)-lysidine synthase | | Authors: | Nakanishi, K, Bonnefond, L, Ishitani, R, Nureki, O. | | Deposit date: | 2009-05-23 | | Release date: | 2009-10-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Structural basis for translational fidelity ensured by transfer RNA lysidine synthetase.
Nature, 461, 2009
|
|
3A1S
 
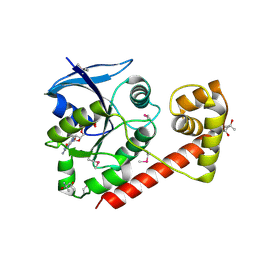 | | Crystal structue of the cytosolic domain of T. maritima FeoB iron iransporter in GDP form I | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Hattori, M, Ishitani, R, Nureki, O. | | Deposit date: | 2009-04-22 | | Release date: | 2009-09-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of novel interactions between the small-GTPase and GDI-like domains in prokaryotic FeoB iron transporter
Structure, 17, 2009
|
|
2ZZT
 
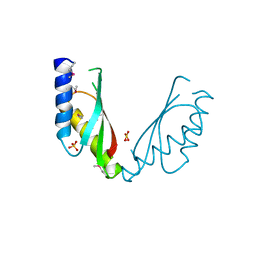 | | Crystal structure of the cytosolic domain of the cation diffusion facilitator family protein | | Descriptor: | Putative uncharacterized protein, SULFATE ION | | Authors: | Higuchi, T, Hattori, M, Tanaka, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2009-02-25 | | Release date: | 2009-08-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.842 Å) | | Cite: | Crystal structure of the cytosolic domain of the cation diffusion facilitator family protein
Proteins, 76, 2009
|
|
2ZZF
 
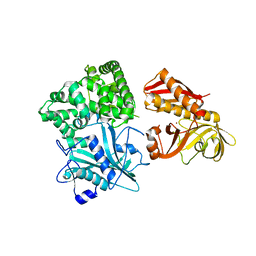 | | Crystal structure of alanyl-tRNA synthetase without oligomerization domain | | Descriptor: | Alanyl-tRNA synthetase, ZINC ION | | Authors: | Sokabe, M, Ose, T, Tokunaga, K, Nakamura, A, Nureki, O, Yao, M, Tanaka, I. | | Deposit date: | 2009-02-10 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of alanyl-tRNA synthetase with editing domain.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3A26
 
 | | Crystal structure of P. horikoshii TYW2 in complex with MeSAdo | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, TETRAETHYLENE GLYCOL, Uncharacterized protein PH0793 | | Authors: | Umitsu, M, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2009-04-28 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of AdoMet-dependent aminocarboxypropyl transfer reaction catalyzed by tRNA-wybutosine synthesizing enzyme, TYW2
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3A4M
 
 | | Crystal structure of archaeal O-phosphoseryl-tRNA(Sec) kinase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, IODIDE ION, ... | | Authors: | Araiso, Y, Ishitani, R, Soll, D, Nureki, O. | | Deposit date: | 2009-07-10 | | Release date: | 2009-10-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Structure of a tRNA-dependent kinase essential for selenocysteine decoding
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2ZZK
 
 | | Crystal structure of tRNA wybutosine synthesizing enzyme TYW4 | | Descriptor: | CITRIC ACID, Leucine carboxyl methyltransferase 2, TETRAETHYLENE GLYCOL | | Authors: | Suzuki, Y, Noma, A, Suzuki, T, Ishitani, R, Nureki, O. | | Deposit date: | 2009-02-17 | | Release date: | 2009-06-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.706 Å) | | Cite: | Structural basis of tRNA modification with CO2 fixation and methylation by wybutosine synthesizing enzyme TYW4.
Nucleic Acids Res., 37, 2009
|
|
3A1W
 
 | |
3A25
 
 | | Crystal structure of P. horikoshii TYW2 in complex with AdoMet | | Descriptor: | S-ADENOSYLMETHIONINE, Uncharacterized protein PH0793 | | Authors: | Umitsu, M, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2009-04-28 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of AdoMet-dependent aminocarboxypropyl transfer reaction catalyzed by tRNA-wybutosine synthesizing enzyme, TYW2
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2ZZG
 
 | | Crystal structure of alanyl-tRNA synthetase in complex with 5''-O-(N-(L-alanyl)-sulfamyoxyl) adenine without oligomerization domain | | Descriptor: | '5'-O-(N-(L-ALANYL)-SULFAMOYL)ADENOSINE, Alanyl-tRNA synthetase, ZINC ION | | Authors: | Sokabe, M, Ose, T, Tokunaga, K, Nakamura, A, Nureki, O, Yao, M, Tanaka, I. | | Deposit date: | 2009-02-10 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structure of alanyl-tRNA synthetase with editing domain.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3A1T
 
 | |
3A1V
 
 | | Crystal structue of the cytosolic domain of T. maritima FeoB iron iransporter in apo form | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Hattori, M, Ishitani, R, Nureki, O. | | Deposit date: | 2009-04-22 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of novel interactions between the small-GTPase and GDI-like domains in prokaryotic FeoB iron transporter
Structure, 17, 2009
|
|
3AL5
 
 | | Crystal structure of Human TYW5 | | Descriptor: | JmjC domain-containing protein C2orf60, MAGNESIUM ION | | Authors: | Kato, M, Araiso, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2010-07-26 | | Release date: | 2010-11-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Crystal structure of a novel JmjC-domain-containing protein, TYW5, involved in tRNA modification.
Nucleic Acids Res., 39, 2011
|
|
3AM1
 
 | | Crystal structure of O-Phosphoseryl-tRNA kinase complexed with anticodon-stem/loop truncated tRNA(Sec) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ASL-truncated tRNA, L-seryl-tRNA(Sec) kinase, ... | | Authors: | Araiso, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2010-08-11 | | Release date: | 2010-12-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | C-terminal domain of archaeal O-phosphoseryl-tRNA kinase displays large-scale motion to bind the 7-bp D-stem of archaeal tRNA(Sec)
Nucleic Acids Res., 39, 2011
|
|
3AQP
 
 | | Crystal structure of SecDF, a translocon-associated membrane protein, from Thermus thrmophilus | | Descriptor: | Probable SecDF protein-export membrane protein | | Authors: | Tsukazaki, T, Mori, H, Echizen, Y, Ishitani, R, Fukai, S, Tanaka, T, Perederina, A, Vassylyev, D.G, Kohno, T, Ito, K, Nureki, O. | | Deposit date: | 2010-11-16 | | Release date: | 2011-05-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure and function of a membrane component SecDF that enhances protein export
Nature, 474, 2011
|
|
2ZJS
 
 | | Crystal Structure of SecYE translocon from Thermus thermophilus with a Fab fragment | | Descriptor: | Fab56 (heavy chain), Fab56 (light chain), Preprotein translocase SecE subunit, ... | | Authors: | Tsukazaki, T, Mori, H, Fukai, S, Ishitani, R, Perederina, A, Vassylyev, D.G, Ito, K, Nureki, O. | | Deposit date: | 2008-03-08 | | Release date: | 2008-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Conformational transition of Sec machinery inferred from bacterial SecYE structures
Nature, 455, 2008
|
|
2ZQP
 
 | |
