8BIN
 
 | | Crystal structure of human Ephrin type-A receptor 2 (EPHA2) Kinase domain in complex with MR21 | | Descriptor: | 1,2-ETHANEDIOL, 8-(4-azanylbutyl)-6-(2-chlorophenyl)-2-(methylamino)pyrido[2,3-d]pyrimidin-7-one, Ephrin type-A receptor 2 | | Authors: | Zhubi, R, Rak, M, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-02 | | Release date: | 2022-11-23 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Back-Pocket Optimization of 2-Aminopyrimidine-Based Macrocycles Leads to Potent EPHA2/GAK Kinase Inhibitors.
J.Med.Chem., 67, 2024
|
|
5EU1
 
 | | CRYSTAL STRUCTURE OF BRD9 IN COMPLEX WITH BI-7273 | | Descriptor: | 4-[4-[(dimethylamino)methyl]-3,5-dimethoxy-phenyl]-2-methyl-2,7-naphthyridin-1-one, BRD9 | | Authors: | Bader, G, Martin, L.M, Steurer, S, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2015-11-18 | | Release date: | 2016-03-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Based Design of an in Vivo Active Selective BRD9 Inhibitor.
J.Med.Chem., 59, 2016
|
|
7Q3N
 
 | | Cryo-EM of the complex between human uromodulin (UMOD)/Tamm-Horsfall protein (THP) and the FimH lectin domain from uropathogenic E. coli | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Type 1 fimbiral adhesin FimH, ... | | Authors: | Jovine, L, Xu, C, Stsiapanava, A, Carroni, M, Tunyasuvunakool, K, Jumper, J, Wu, B. | | Deposit date: | 2021-10-28 | | Release date: | 2022-03-16 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8BZJ
 
 | | Human MST3 (STK24) kinase in complex with inhibitor MRLW5 | | Descriptor: | 1,2-ETHANEDIOL, 8-(4-azanylbutyl)-6-[2-chloranyl-4-(6-methylpyridin-2-yl)phenyl]-2-(methylamino)pyrido[2,3-d]pyrimidin-7-one, Serine/threonine-protein kinase 24 | | Authors: | Balourdas, D.I, Rak, M, Tesch, R, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-12-14 | | Release date: | 2023-01-18 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Development of Selective Pyrido[2,3- d ]pyrimidin-7(8 H )-one-Based Mammalian STE20-Like (MST3/4) Kinase Inhibitors.
J.Med.Chem., 67, 2024
|
|
5MW2
 
 | | CRYSTAL STRUCTURE OF BCL-6 BTB-domain with BI-3802 | | Descriptor: | 2-[6-[[5-chloranyl-2-[(3~{S},5~{R})-3,5-dimethylpiperidin-1-yl]pyrimidin-4-yl]amino]-1-methyl-2-oxidanylidene-quinolin-3-yl]oxy-~{N}-methyl-ethanamide, B-cell lymphoma 6 protein | | Authors: | Bader, G, Flotzinger, G, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2017-01-18 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Chemically Induced Degradation of the Oncogenic Transcription Factor BCL6.
Cell Rep, 20, 2017
|
|
5MW6
 
 | | Crystal structure of the BCL6 BTB-domain with compound 1 | | Descriptor: | 5-chloranyl-~{N}-(4-chlorophenyl)-2-(3,5-dimethylpyrazol-1-yl)pyrimidin-4-amine, B-cell lymphoma 6 protein | | Authors: | Davies, D.R, Kessler, D. | | Deposit date: | 2017-01-18 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Chemically Induced Degradation of the Oncogenic Transcription Factor BCL6.
Cell Rep, 20, 2017
|
|
5MWD
 
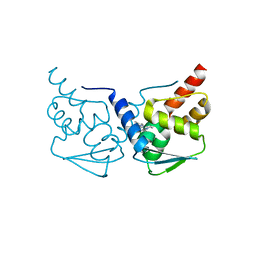 | | Crystal structure of the BCL6 BTB-domain with compound 2 | | Descriptor: | 5-[[5-chloranyl-2-(3,5-dimethylpyrazol-1-yl)pyrimidin-4-yl]amino]-1,3-dihydroindol-2-one, B-cell lymphoma 6 protein | | Authors: | Bader, G, Flotzinger, G, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2017-01-18 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Chemically Induced Degradation of the Oncogenic Transcription Factor BCL6.
Cell Rep, 20, 2017
|
|
4XZ2
 
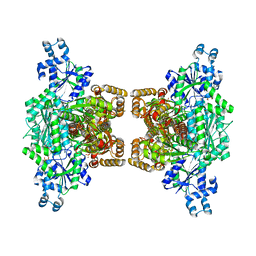 | | Human platelet phosphofructokinase in an R-state in complex with ADP and F6P, crystal form I | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 6-O-phosphono-beta-D-fructofuranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kloos, M, Strater, N. | | Deposit date: | 2015-02-03 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Crystal structure of human platelet phosphofructokinase-1 locked in an activated conformation.
Biochem.J., 469, 2015
|
|
5E4D
 
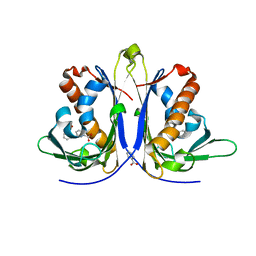 | |
5E46
 
 | |
5E4B
 
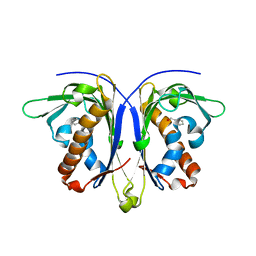 | |
9BCG
 
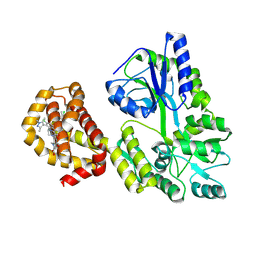 | | Myeloid cell leukemia-1 (Mcl-1) complexed with compound | | Descriptor: | 7-[(4R,5S,6P)-7-chloro-10-[3-(4-chloro-3,5-dimethylphenoxy)propyl]-4-methyl-1-oxo-6-(1,3,5-trimethyl-1H-pyrazol-4-yl)-3,4-dihydropyrazino[1,2-a]indol-2(1H)-yl]-4,5-dimethoxy-1-methyl-1H-indole-2-carboxylic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Zhao, B, Fesik, S.W. | | Deposit date: | 2024-04-09 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Discovery of a Myeloid Cell Leukemia 1 (Mcl-1) Inhibitor That Demonstrates Potent In Vivo Activities in Mouse Models of Hematological and Solid Tumors.
J.Med.Chem., 67, 2024
|
|
8QLR
 
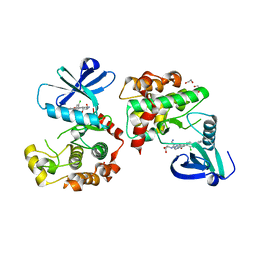 | | Human MST3 (STK24) kinase in complex with inhibitor MR24 | | Descriptor: | 1,2-ETHANEDIOL, 8-(4-azanylbutyl)-2-[1,3-bis(oxidanyl)propan-2-ylamino]-6-[2-chloranyl-4-(6-methylpyridin-2-yl)phenyl]pyrido[2,3-d]pyrimidin-7-one, Serine/threonine-protein kinase 24 | | Authors: | Balourdas, D.I, Rak, M, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-09-20 | | Release date: | 2023-11-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Development of Selective Pyrido[2,3- d ]pyrimidin-7(8 H )-one-Based Mammalian STE20-Like (MST3/4) Kinase Inhibitors.
J.Med.Chem., 67, 2024
|
|
8QLT
 
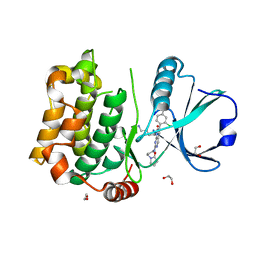 | | Human MST3 (STK24) kinase in complex with inhibitor MR30 | | Descriptor: | 1,2-ETHANEDIOL, 8-(4-azanylbutyl)-6-[2-chloranyl-4-(6-methylpyridin-2-yl)phenyl]-2-[3-(2-oxidanylidenepyrrolidin-1-yl)propylamino]pyrido[2,3-d]pyrimidin-7-one, Serine/threonine-protein kinase 24 | | Authors: | Balourdas, D.I, Rak, M, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-09-20 | | Release date: | 2023-11-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Development of Selective Pyrido[2,3- d ]pyrimidin-7(8 H )-one-Based Mammalian STE20-Like (MST3/4) Kinase Inhibitors.
J.Med.Chem., 67, 2024
|
|
8QLS
 
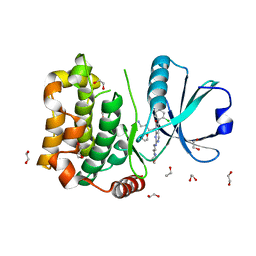 | | Human MST3 (STK24) kinase in complex with inhibitor MR26 | | Descriptor: | 1,2-ETHANEDIOL, 8-(4-azanylbutyl)-6-[2-chloranyl-4-(6-methylpyridin-2-yl)phenyl]-2-(3-morpholin-4-ylpropylamino)pyrido[2,3-d]pyrimidin-7-one, Serine/threonine-protein kinase 24 | | Authors: | Balourdas, D.I, Rak, M, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-09-20 | | Release date: | 2023-11-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Development of Selective Pyrido[2,3- d ]pyrimidin-7(8 H )-one-Based Mammalian STE20-Like (MST3/4) Kinase Inhibitors.
J.Med.Chem., 67, 2024
|
|
7UQ2
 
 | | Vs.4 from T4 phage in complex with cGAMP | | Descriptor: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, CALCIUM ION, Vs.4 | | Authors: | Jenson, J.M, Chen, Z.J. | | Deposit date: | 2022-04-18 | | Release date: | 2023-02-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ubiquitin-like conjugation by bacterial cGAS enhances anti-phage defence.
Nature, 616, 2023
|
|
7PFP
 
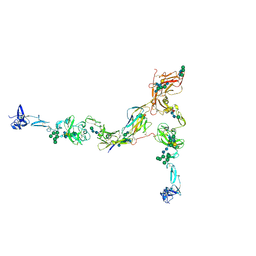 | | Full-length cryo-EM structure of the native human uromodulin (UMOD)/Tamm-Horsfall protein (THP) filament | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Uromodulin, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jovine, L, Xu, C, Stsiapanava, A, Carroni, M, Tunyasuvunakool, K, Jumper, J, Wu, B. | | Deposit date: | 2021-08-11 | | Release date: | 2022-03-16 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8BIO
 
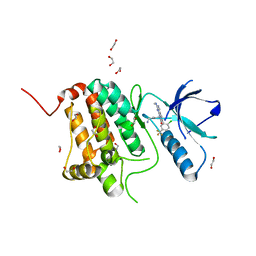 | | Crystal structure of human Ephrin type-A receptor 2 (EPHA2) Kinase domain in complex with MRAL5 | | Descriptor: | 1,2-ETHANEDIOL, 8-(4-azanylbutyl)-6-[2-chloranyl-5-(trifluoromethyl)phenyl]-2-(methylamino)pyrido[2,3-d]pyrimidin-7-one, Ephrin type-A receptor 2 | | Authors: | Zhubi, R, Rak, M, Lucic, A, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-02 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Shifting the selectivity of pyrido[2,3-d]pyrimidin-7(8H)-one inhibitors towards the salt-inducible kinase (SIK) subfamily.
Eur.J.Med.Chem., 254, 2023
|
|
1AZH
 
 | |
1AZJ
 
 | |
1AZK
 
 | |
8BZI
 
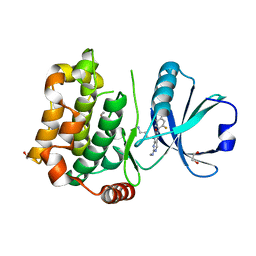 | | Human MST3 (STK24) kinase in complex with inhibitor MR39 | | Descriptor: | 1,2-ETHANEDIOL, 8-(4-azanylbutyl)-6-[2,5-bis(fluoranyl)-4-(6-methylpyridin-2-yl)phenyl]-2-(methylamino)pyrido[2,3-d]pyrimidin-7-one, Serine/threonine-protein kinase 24 | | Authors: | Balourdas, D.I, Rak, M, Tesch, R, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-12-14 | | Release date: | 2023-01-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Shifting the selectivity of pyrido[2,3-d]pyrimidin-7(8H)-one inhibitors towards the salt-inducible kinase (SIK) subfamily.
Eur.J.Med.Chem., 254, 2023
|
|
4CML
 
 | | Crystal Structure of INPP5B in complex with Phosphatidylinositol 3,4- bisphosphate | | Descriptor: | 1,2-dioctanoyl phosphatidyl epi-inositol (3,4)-bisphosphate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Tresaugues, L, Arrowsmith, C.H, Berglund, H, Bountra, C, Edwards, A.M, Ekblad, T, Flodin, S, Graslund, S, Karlberg, T, Moche, M, Nyman, T, Schuler, H, Silvander, C, Thorsell, A.G, Weigelt, J, Welin, M, Nordlund, P. | | Deposit date: | 2014-01-16 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Phosphoinositide Substrate Recognition, Catalysis, and Membrane Interactions in Human Inositol Polyphosphate 5-Phosphatases.
Structure, 22, 2014
|
|
1AZ6
 
 | |
4CMN
 
 | | Crystal structure of OCRL in complex with a phosphate ion | | Descriptor: | GLYCEROL, INOSITOL POLYPHOSPHATE 5-PHOSPHATASE OCRL-1, MAGNESIUM ION, ... | | Authors: | Tresaugues, L, Moche, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Edwards, A.M, Ekblad, T, Flodin, S, Graslund, S, Karlberg, T, Nyman, T, Schuler, H, Silvander, C, Thorsell, A.G, Weigelt, J, Welin, M, Nordlund, P. | | Deposit date: | 2014-01-16 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Structural Basis for Phosphoinositide Substrate Recognition, Catalysis, and Membrane Interactions in Human Inositol Polyphosphate 5-Phosphatases.
Structure, 22, 2014
|
|
