8RLE
 
 | | SPNS2:sfGFP hetero dimer assembled by Di-Gluebody - sfGFP local refinement | | Descriptor: | Gluebody GbC4, Gluebody GbEnhancer, Green fluorescent protein | | Authors: | Yi, G, Ye, M, Mamalis, D, Sauer, D.B, von Delft, F, Davis, B.G, Gilbert, R.J.C. | | Deposit date: | 2024-01-02 | | Release date: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Di-Gluebodies: Rigid modular nanobody protein assemblies enabling simultaneous determination of high-resolution cryo-EM structures
To Be Published
|
|
6MGP
 
 | | Structure of human 4-1BB / 4-1BBL complex | | Descriptor: | ACETATE ION, GLYCEROL, Tumor necrosis factor ligand superfamily member 9, ... | | Authors: | Kimberlin, C.R, Chin, S.M, Roe-Zurz, Z, Xu, A, Yang, Y. | | Deposit date: | 2018-09-14 | | Release date: | 2018-11-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structure of the 4-1BB/4-1BBL complex and distinct binding and functional properties of utomilumab and urelumab
Nat Commun, 9, 2018
|
|
6MGE
 
 | | Structure of human 4-1BBL | | Descriptor: | GLYCEROL, PHOSPHATE ION, Tumor necrosis factor ligand superfamily member 9 | | Authors: | Kimberlin, C.R, Chin, S.M, Roe-Zurz, Z, Xu, A, Yang, Y. | | Deposit date: | 2018-09-13 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of the 4-1BB/4-1BBL complex and distinct binding and functional properties of utomilumab and urelumab.
Nat Commun, 9, 2018
|
|
6MHR
 
 | | Structure of the human 4-1BB / Urelumab Fab complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, MALONATE ION, ... | | Authors: | Kimberlin, C.R, Chin, S.M, Roe-Zurz, Z, Xu, A, Yang, Y. | | Deposit date: | 2018-09-18 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the 4-1BB/4-1BBL complex and distinct binding and functional properties of utomilumab and urelumab.
Nat Commun, 9, 2018
|
|
6MI2
 
 | | Structure of the human 4-1BB / Utomilumab Fab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, SULFATE ION, ... | | Authors: | Kimberlin, C.R, Chin, S.M, Roe-Zurz, Z, Xu, A, Yang, Y. | | Deposit date: | 2018-09-19 | | Release date: | 2018-11-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structure of the 4-1BB/4-1BBL complex and distinct binding and functional properties of utomilumab and urelumab.
Nat Commun, 9, 2018
|
|
8TH1
 
 | |
8TH7
 
 | |
8TH6
 
 | | Crystal Structure of the G3BP1 NTF2-like domain bound to USP10 peptide | | Descriptor: | 1,2-ETHANEDIOL, Ras GTPase-activating protein-binding protein 1, Ubiquitin carboxyl-terminal hydrolase 10 | | Authors: | Hughes, M.P, Taylor, J.P, Yang, Z. | | Deposit date: | 2023-07-14 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Interaction between host G3BP and viral nucleocapsid protein regulates SARS-CoV-2 replication and pathogenicity.
Cell Rep, 43, 2024
|
|
8TH5
 
 | |
6O1D
 
 | |
6NCJ
 
 | | Structure of HIV-1 Integrase with potent 5,6,7,8-Tetrahydro-1,6-naphthyridine Derivatives Allosteric Site Inhibitors | | Descriptor: | (2~{S})-2-[4-(8-fluoranyl-5-methyl-3,4-dihydro-2~{H}-chromen-6-yl)-2-methyl-6-[[(1~{S},2~{R})-2-phenylcyclopropyl]methyl]-7,8-dihydro-5~{H}-1,6-naphthyridin-3-yl]-2-[(2-methylpropan-2-yl)oxy]ethanoic acid, 1,2-ETHANEDIOL, Integrase, ... | | Authors: | Nolte, R.T. | | Deposit date: | 2018-12-11 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 5,6,7,8-Tetrahydro-1,6-naphthyridine Derivatives as Potent HIV-1-Integrase-Allosteric-Site Inhibitors.
J. Med. Chem., 62, 2019
|
|
6OKO
 
 | |
6NO7
 
 | | Crystal Structure of the full-length wild-type PKA RIa Holoenzyme | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Lu, T, Wu, J, Taylor, S.S. | | Deposit date: | 2019-01-15 | | Release date: | 2019-07-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Two PKA RI alpha holoenzyme states define ATP as an isoform-specific orthosteric inhibitor that competes with the allosteric activator, cAMP.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7YI3
 
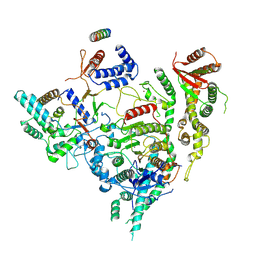 | | Cryo-EM structure of Rpd3S in close-state Rpd3S-NCP complex | | Descriptor: | Chromatin modification-related protein EAF3, Histone deacetylase RPD3, Transcriptional regulatory protein RCO1, ... | | Authors: | Li, H.T, Yan, C.Y, Guan, H.P, Wang, P. | | Deposit date: | 2022-07-14 | | Release date: | 2023-06-14 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Diverse modes of H3K36me3-guided nucleosomal deacetylation by Rpd3S.
Nature, 620, 2023
|
|
7YI2
 
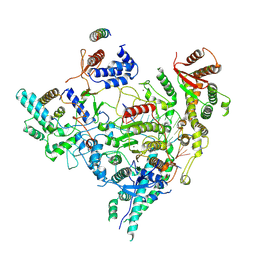 | | Cryo-EM structure of Rpd3S in loose-state Rpd3S-NCP complex | | Descriptor: | Chromatin modification-related protein EAF3, Histone deacetylase RPD3, Transcriptional regulatory protein RCO1, ... | | Authors: | Li, H.T, Yan, C.Y, Guan, H.P, Wang, P. | | Deposit date: | 2022-07-14 | | Release date: | 2023-06-14 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Diverse modes of H3K36me3-guided nucleosomal deacetylation by Rpd3S.
Nature, 620, 2023
|
|
7YI1
 
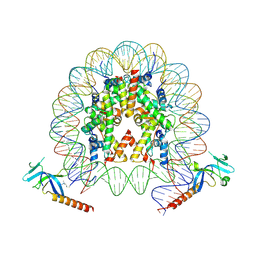 | | Cryo-EM structure of Eaf3 CHD bound to H3K36me3 nucleosome | | Descriptor: | Chromatin modification-related protein EAF3, Histone H2A, Histone H2B 1.1, ... | | Authors: | Li, H.T, Yan, C.Y, Guan, H.P, Wang, P. | | Deposit date: | 2022-07-14 | | Release date: | 2023-06-14 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Diverse modes of H3K36me3-guided nucleosomal deacetylation by Rpd3S.
Nature, 620, 2023
|
|
7YI0
 
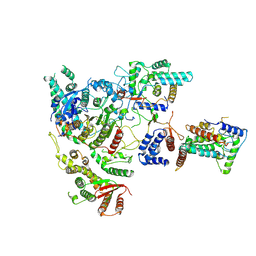 | | Cryo-EM structure of Rpd3S complex | | Descriptor: | Chromatin modification-related protein EAF3, Histone deacetylase RPD3, Transcriptional regulatory protein RCO1, ... | | Authors: | Li, H.T, Yan, C.Y, Guan, H.P, Wang, P. | | Deposit date: | 2022-07-14 | | Release date: | 2023-06-14 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Diverse modes of H3K36me3-guided nucleosomal deacetylation by Rpd3S.
Nature, 620, 2023
|
|
5UPW
 
 | |
8BSM
 
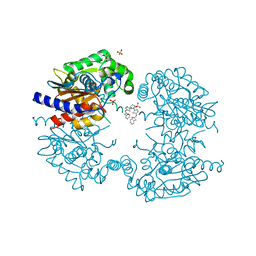 | | Human GLS in complex with compound 18 | | Descriptor: | 2-phenyl-~{N}-[5-[[(3~{R})-1-[5-(2-phenylethanoylamino)-1,3,4-thiadiazol-2-yl]pyrrolidin-3-yl]amino]-1,3,4-thiadiazol-2-yl]ethanamide, Glutaminase kidney isoform, mitochondrial 65 kDa chain, ... | | Authors: | Debreczeni, J.E. | | Deposit date: | 2022-11-25 | | Release date: | 2023-01-18 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.782 Å) | | Cite: | Discovery of a Thiadiazole-Pyridazine-Based Allosteric Glutaminase 1 Inhibitor Series That Demonstrates Oral Bioavailability and Activity in Tumor Xenograft Models.
J Med Chem, 62, 2019
|
|
8BSK
 
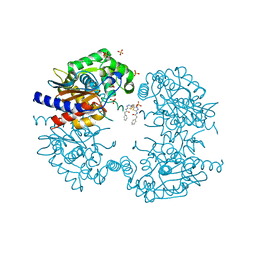 | | Human GLS in complex with compound 3 | | Descriptor: | Glutaminase kidney isoform, mitochondrial 65 kDa chain, N,N'-[sulfanediylbis(ethane-2,1-diyl-1,3,4-thiadiazole-5,2-diyl)]bis(2-phenylacetamide), ... | | Authors: | Debreczeni, J.E. | | Deposit date: | 2022-11-25 | | Release date: | 2023-01-18 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of a Thiadiazole-Pyridazine-Based Allosteric Glutaminase 1 Inhibitor Series That Demonstrates Oral Bioavailability and Activity in Tumor Xenograft Models.
J Med Chem, 62, 2019
|
|
8BSL
 
 | | Human GLS in complex with compound 12 | | Descriptor: | Glutaminase kidney isoform, mitochondrial, ~{N}-[5-[[(3~{R})-1-(5-azanyl-1,3,4-thiadiazol-2-yl)pyrrolidin-3-yl]amino]-1,3,4-thiadiazol-2-yl]-2-phenyl-ethanamide | | Authors: | Debreczeni, J.E. | | Deposit date: | 2022-11-25 | | Release date: | 2023-01-18 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Discovery of a Thiadiazole-Pyridazine-Based Allosteric Glutaminase 1 Inhibitor Series That Demonstrates Oral Bioavailability and Activity in Tumor Xenograft Models.
J Med Chem, 62, 2019
|
|
8BSN
 
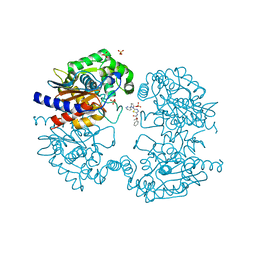 | | Human GLS in complex with compound 27 | | Descriptor: | (2~{S})-2-methoxy-2-phenyl-~{N}-[5-[[(3~{R})-1-pyridazin-3-ylpyrrolidin-3-yl]amino]-1,3,4-thiadiazol-2-yl]ethanamide, Glutaminase kidney isoform, mitochondrial 65 kDa chain, ... | | Authors: | Debreczeni, J.E. | | Deposit date: | 2022-11-25 | | Release date: | 2023-01-18 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | Discovery of a Thiadiazole-Pyridazine-Based Allosteric Glutaminase 1 Inhibitor Series That Demonstrates Oral Bioavailability and Activity in Tumor Xenograft Models.
J Med Chem, 62, 2019
|
|
7YI5
 
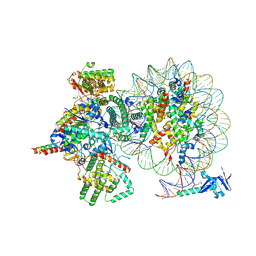 | | Cryo-EM structure of Rpd3S complex bound to H3K36me3 nucleosome in loose state | | Descriptor: | Chromatin modification-related protein EAF3, Histone H2A, Histone H2B 1.1, ... | | Authors: | Li, H.T, Yan, C.Y, Guan, H.P, Wang, P. | | Deposit date: | 2022-07-14 | | Release date: | 2023-06-14 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Diverse modes of H3K36me3-guided nucleosomal deacetylation by Rpd3S.
Nature, 620, 2023
|
|
7YI4
 
 | | Cryo-EM structure of Rpd3S complex bound to H3K36me3 nucleosome in close state | | Descriptor: | Chromatin modification-related protein EAF3, Histone H2A, Histone H2B 1.1, ... | | Authors: | Li, H.T, Yan, C.Y, Guan, H.P, Wang, P. | | Deposit date: | 2022-07-14 | | Release date: | 2023-06-14 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Diverse modes of H3K36me3-guided nucleosomal deacetylation by Rpd3S.
Nature, 620, 2023
|
|
5WCU
 
 | |
