1G6R
 
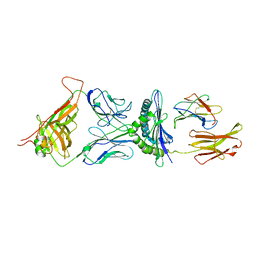 | | A FUNCTIONAL HOT SPOT FOR ANTIGEN RECOGNITION IN A SUPERAGONIST TCR/MHC COMPLEX | | Descriptor: | ALPHA T CELL RECEPTOR, BETA T CELL RECEPTOR, BETA-2 MICROGLOBULIN, ... | | Authors: | Degano, M, Garcia, K.C, Apostolopoulos, V, Rudolph, M.G, Teyton, L, Wilson, I.A. | | Deposit date: | 2000-11-07 | | Release date: | 2000-11-15 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A functional hot spot for antigen recognition in a superagonist TCR/MHC complex.
Immunity, 12, 2000
|
|
1GND
 
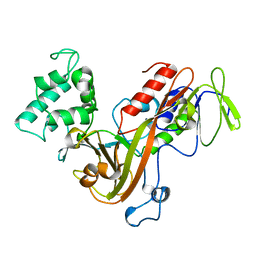 | | GUANINE NUCLEOTIDE DISSOCIATION INHIBITOR, ALPHA-ISOFORM | | Descriptor: | GUANINE NUCLEOTIDE DISSOCIATION INHIBITOR | | Authors: | Schalk, I, Zeng, K, Wu, S.-K, Stura, E.A, Metteson, J, Huang, M, Tandon, A, Wilson, I.A, Balch, W.E. | | Deposit date: | 1996-07-10 | | Release date: | 1997-02-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure and mutational analysis of Rab GDP-dissociation inhibitor.
Nature, 381, 1996
|
|
1EBA
 
 | |
1GRC
 
 | |
1HIM
 
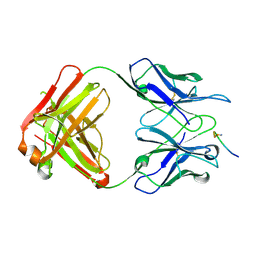 | |
1HIL
 
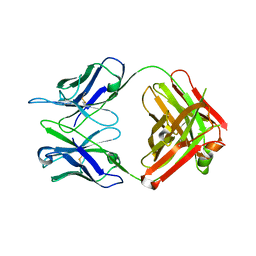 | |
1A3L
 
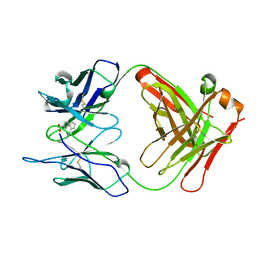 | |
1AXT
 
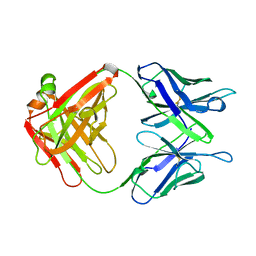 | |
2O5Y
 
 | |
1AHW
 
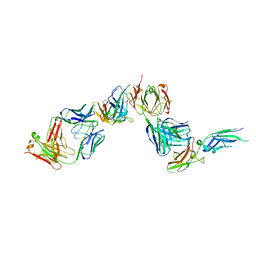 | | A COMPLEX OF EXTRACELLULAR DOMAIN OF TISSUE FACTOR WITH AN INHIBITORY FAB (5G9) | | Descriptor: | IMMUNOGLOBULIN FAB 5G9 (HEAVY CHAIN), IMMUNOGLOBULIN FAB 5G9 (LIGHT CHAIN), TISSUE FACTOR | | Authors: | Huang, M, Syed, R, Stura, E.A, Stone, M.J, Stefanko, R.S, Ruf, W, Edgington, T.S, Wilson, I.A. | | Deposit date: | 1997-04-10 | | Release date: | 1998-02-25 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The mechanism of an inhibitory antibody on TF-initiated blood coagulation revealed by the crystal structures of human tissue factor, Fab 5G9 and TF.5G9 complex.
J.Mol.Biol., 275, 1998
|
|
2OJZ
 
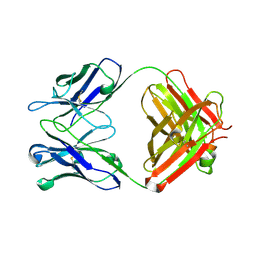 | | Anti-DNA antibody ED10 | | Descriptor: | Fab ED10 heavy chain, Fab ED10 light chain | | Authors: | Stanfield, R.L, Sanguineti, S, Wilson, I.A, de Prat-Gay, G. | | Deposit date: | 2007-01-15 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Specific recognition of a DNA immunogen by its elicited antibody
J.Mol.Biol., 370, 2007
|
|
1ACY
 
 | |
1C5B
 
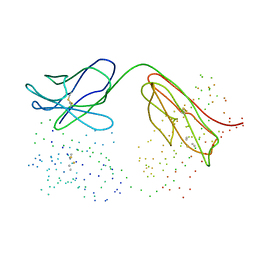 | | DECARBOXYLASE CATALYTIC ANTIBODY 21D8 UNLIGANDED FORM | | Descriptor: | CHIMERIC DECARBOXYLASE ANTIBODY 21D8 | | Authors: | Hotta, K, Wilson, I.A. | | Deposit date: | 1999-11-08 | | Release date: | 2000-10-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Catalysis of decarboxylation by a preorganized heterogeneous microenvironment: crystal structures of abzyme 21D8.
J.Mol.Biol., 302, 2000
|
|
2O5X
 
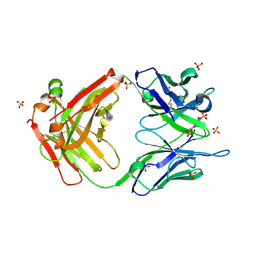 | |
1C5C
 
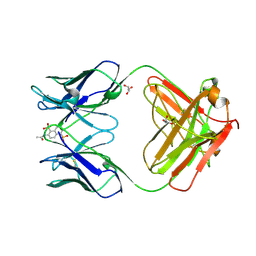 | | DECARBOXYLASE CATALYTIC ANTIBODY 21D8-HAPTEN COMPLEX | | Descriptor: | 2-ACETYLAMINO-NAPTHALENE-1,5-DISULFONIC ACID, CHIMERIC DECARBOXYLASE ANTIBODY 21D8, GLYCEROL | | Authors: | Hotta, K, Wilson, I.A. | | Deposit date: | 1999-11-09 | | Release date: | 2000-10-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Catalysis of decarboxylation by a preorganized heterogeneous microenvironment: crystal structures of abzyme 21D8.
J.Mol.Biol., 302, 2000
|
|
1C16
 
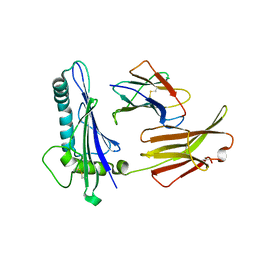 | | CRYSTAL STRUCTURE ANALYSIS OF THE GAMMA/DELTA T CELL LIGAND T22 | | Descriptor: | MHC-LIKE PROTEIN T22, PROTEIN (BETA-2-MICROGLOBULIN) | | Authors: | Wingren, C, Crowley, M.P, Degano, M, Chien, Y, Wilson, I.A. | | Deposit date: | 1999-07-20 | | Release date: | 2000-01-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of a gammadelta T cell receptor ligand T22: a truncated MHC-like fold.
Science, 287, 2000
|
|
1AI1
 
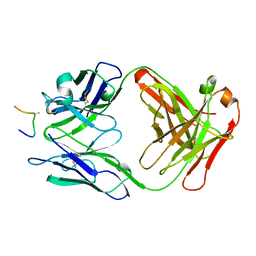 | | HIV-1 V3 LOOP MIMIC | | Descriptor: | AIB142, IGG1-KAPPA 59.1 FAB (HEAVY CHAIN), IGG1-KAPPA 59.1 FAB (LIGHT CHAIN) | | Authors: | Ghiara, J.B, Wilson, I.A. | | Deposit date: | 1996-11-06 | | Release date: | 1997-05-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of a constrained peptide mimic of the HIV-1 V3 loop neutralization site.
J.Mol.Biol., 266, 1997
|
|
2OQJ
 
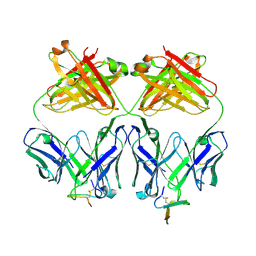 | | Crystal structure analysis of Fab 2G12 in complex with peptide 2G12.1 | | Descriptor: | Fab 2G12 heavy chain, Fab 2G12 light chain, peptide 2G12.1 (ACPPSHVLDMRSGTCLAAEGK) | | Authors: | Calarese, D.A, Stanfield, R.L, Menendez, A, Scott, J.K, Wilson, I.A. | | Deposit date: | 2007-01-31 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A peptide inhibitor of HIV-1 neutralizing antibody 2G12 is not a structural mimic of the natural carbohydrate epitope on gp120.
Faseb J., 22, 2008
|
|
2OP4
 
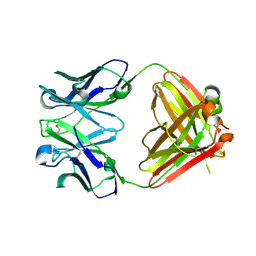 | | Crystal Structure of Quorum-Quenching Antibody 1G9 | | Descriptor: | 1,2-ETHANEDIOL, Murine Antibody Fab RS2-1G9 IGG1 Heavy Chain, Murine Antibody Fab RS2-1G9 Lambda Light Chain | | Authors: | Kirchdoerfer, R.N, Debler, E.W, Wilson, I.A. | | Deposit date: | 2007-01-26 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structures of a Quorum-quenching Antibody.
J.Mol.Biol., 368, 2007
|
|
2OK0
 
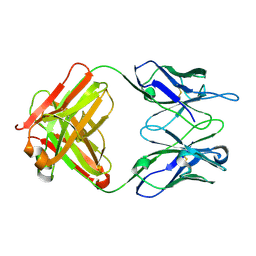 | | Fab ED10-DNA complex | | Descriptor: | 5'-D(*TP*C)-3', Fab ED10 heavy chain, Fab ED10 light chain | | Authors: | Stanfield, R.L, Sanguineti, S, Wilson, I.A, de Prat-Gay, G. | | Deposit date: | 2007-01-15 | | Release date: | 2007-06-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Specific recognition of a DNA immunogen by its elicited antibody
J.Mol.Biol., 370, 2007
|
|
2O5Z
 
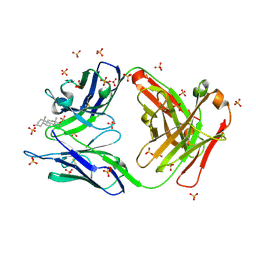 | |
1D5T
 
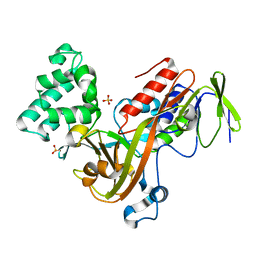 | | GUANINE NUCLEOTIDE DISSOCIATION INHIBITOR, ALPHA-ISOFORM | | Descriptor: | GUANINE NUCLEOTIDE DISSOCIATION INHIBITOR, SULFATE ION | | Authors: | Peng, L, Zeng, K, Heine, A, Moyer, B, Greasley, S.E, Kuhn, P, Balch, W.E, Wilson, I.A. | | Deposit date: | 1999-10-11 | | Release date: | 2000-10-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | A new functional domain of guanine nucleotide dissociation inhibitor (alpha-GDI) involved in Rab recycling.
Traffic, 1, 2000
|
|
2NTF
 
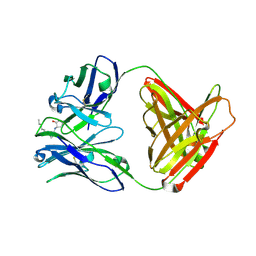 | |
1C1E
 
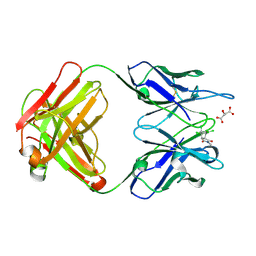 | | CRYSTAL STRUCTURE OF A DIELS-ALDERASE CATALYTIC ANTIBODY 1E9 IN COMPLEX WITH ITS HAPTEN | | Descriptor: | 1,7,8,9,10,10-HEXACHLORO-4-METHYL-4-AZA-TRICYCLO[5.2.1.0(2,6)]DEC-8-ENE-3,5-DIONE, CATALYTIC ANTIBODY 1E9 (HEAVY CHAIN), CATALYTIC ANTIBODY 1E9 (LIGHT CHAIN), ... | | Authors: | Xu, J, Wilson, I.A. | | Deposit date: | 1999-07-22 | | Release date: | 2000-03-01 | | Last modified: | 2013-01-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolution of shape complementarity and catalytic efficiency from a primordial antibody template.
Science, 286, 1999
|
|
2PRT
 
 | | Structure of the Wilms Tumor Suppressor Protein Zinc Finger Domain Bound to DNA | | Descriptor: | DNA (5'-D(*CP*AP*GP*AP*CP*GP*CP*CP*CP*CP*CP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*GP*GP*GP*GP*GP*CP*GP*TP*CP*TP*G)-3'), Wilms tumor 1, ... | | Authors: | Stoll, R, Lee, B.M, Debler, E.W, Laity, J.H, Wilson, I.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2007-05-04 | | Release date: | 2008-03-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structure of the Wilms tumor suppressor protein zinc finger domain bound to DNA
J.Mol.Biol., 372, 2007
|
|
