6JSC
 
 | |
6JSD
 
 | |
6JSB
 
 | |
2ZUR
 
 | | Crystal Structure of Rh(nbd)/apo-Fr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, Ferritin light chain, ... | | Authors: | Abe, S, Hirata, K, Ueno, T, Shimizu, N, Yamamoto, M, Takata, M, Watanabe, Y. | | Deposit date: | 2008-10-28 | | Release date: | 2009-06-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Polymerization of phenylacetylene by rhodium complexes within a discrete space of apo-ferritin
J.Am.Chem.Soc., 131, 2009
|
|
5URM
 
 | | Crystal structure of human BRR2 in complex with T-1206548 | | Descriptor: | 3-(5-{[(2R)-5-amino-2-cyclohexyl-7-oxo-2,3-dihydro-7H-[1,3,4]thiadiazolo[3,2-a]pyrimidin-6-yl]methyl}furan-2-yl)benzoic acid, U5 small nuclear ribonucleoprotein 200 kDa helicase | | Authors: | Klein, M.G, Tjhen, R, Qin, L. | | Deposit date: | 2017-02-11 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of Allosteric Inhibitors Targeting the Spliceosomal RNA Helicase Brr2.
J. Med. Chem., 60, 2017
|
|
5URJ
 
 | | Crystal structure of human BRR2 in complex with T-3905516 | | Descriptor: | 6-benzyl-3-[(2R)-2-(3-fluoropyridin-2-yl)-6-methyl-3,4-dihydro-2H-1-benzopyran-7-yl]-4,6-dihydropyrido[4,3-d]pyrimidine-2,7(3H,8H)-dione, GLYCEROL, U5 small nuclear ribonucleoprotein 200 kDa helicase | | Authors: | Klein, M.G, Tjhen, R, Qin, L. | | Deposit date: | 2017-02-10 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Discovery of Allosteric Inhibitors Targeting the Spliceosomal RNA Helicase Brr2.
J. Med. Chem., 60, 2017
|
|
5URK
 
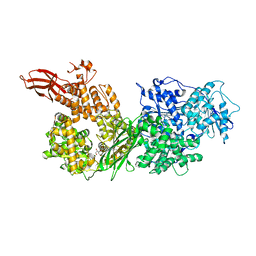 | | Crystal structure of human BRR2 in complex with T-3935799 | | Descriptor: | 6-benzyl-3-[3-(benzyloxy)phenyl]-4,6-dihydropyrido[4,3-d]pyrimidine-2,7(1H,3H)-dione, GLYCEROL, U5 small nuclear ribonucleoprotein 200 kDa helicase | | Authors: | Qin, L, Tjhen, R, Klein, M.G. | | Deposit date: | 2017-02-10 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Discovery of Allosteric Inhibitors Targeting the Spliceosomal RNA Helicase Brr2.
J. Med. Chem., 60, 2017
|
|
3A4P
 
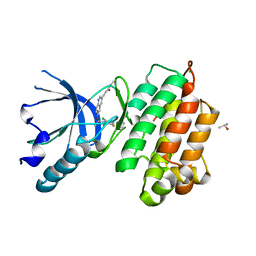 | | human c-MET kinase domain complexed with 6-benzyloxyquinoline inhibitor | | Descriptor: | (2E)-3-{6-[(1R)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]quinolin-3-yl}-N-methylprop-2-enamide, CHLORIDE ION, Hepatocyte growth factor receptor, ... | | Authors: | Fukami, T.A, Kadono, S, Yamamuro, M, Matsuura, T. | | Deposit date: | 2009-07-13 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Discovery of 6-benzyloxyquinolines as c-Met selective kinase inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
5B50
 
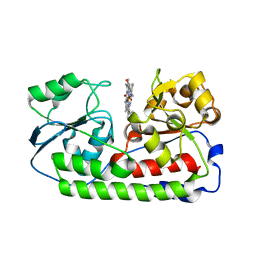 | | Crystal structure of heme binding protein HmuT Y240A | | Descriptor: | ABC-type transporter, periplasmic component, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Muraki, N, Aono, S. | | Deposit date: | 2016-04-20 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Characterization of Heme Environmental Mutants of CgHmuT that Shuttles Heme Molecules to Heme Transporters
Int J Mol Sci, 17, 2016
|
|
7WRR
 
 | |
5B51
 
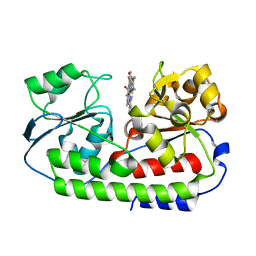 | | Crystal structure of heme binding protein HmuT R242A mutant | | Descriptor: | ABC-type transporter, periplasmic component, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Muraki, N, Aono, S. | | Deposit date: | 2016-04-20 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Characterization of Heme Environmental Mutants of CgHmuT that Shuttles Heme Molecules to Heme Transporters
Int J Mol Sci, 17, 2016
|
|
5B4Z
 
 | | Crystal structure of heme binding protein HmuT H141A mutant | | Descriptor: | ABC-type transporter, periplasmic component, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Muraki, N, Aono, S. | | Deposit date: | 2016-04-20 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Characterization of Heme Environmental Mutants of CgHmuT that Shuttles Heme Molecules to Heme Transporters
Int J Mol Sci, 17, 2016
|
|
7WRT
 
 | |
5DE0
 
 | | Dye-decolorizing protein from V. cholerae | | Descriptor: | Deferrochelatase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Uchida, T, Sasaki, M, Tanaka, Y, Yao, M. | | Deposit date: | 2015-08-25 | | Release date: | 2015-10-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | A Dye-Decolorizing Peroxidase from Vibrio cholerae.
Biochemistry, 54, 2015
|
|
2V6L
 
 | | Molecular Model of a Type III Secretion System Needle | | Descriptor: | MXIH | | Authors: | Deane, J.E, Roversi, P, Cordes, F.S, Johnson, S, Kenjale, R, Daniell, S, Booy, F, Picking, W.L, Picking, W.D, Blocker, A.J, Lea, S.M. | | Deposit date: | 2007-07-19 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (16 Å) | | Cite: | Molecular model of a type III secretion system needle: Implications for host-cell sensing.
Proc. Natl. Acad. Sci. U.S.A., 103, 2006
|
|
5XCO
 
 | |
2D5K
 
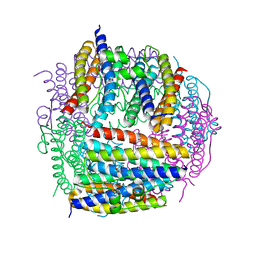 | | Crystal structure of Dps from Staphylococcus aureus | | Descriptor: | Dps family protein, GLYCEROL | | Authors: | Tanaka, Y, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2005-11-02 | | Release date: | 2006-10-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Nucleoid compaction by MrgA(Asp56Ala/Glu60Ala) does not contribute to staphylococcal cell survival against oxidative stress and phagocytic killing by macrophages
FEMS Microbiol. Lett., 360, 2014
|
|
3VYL
 
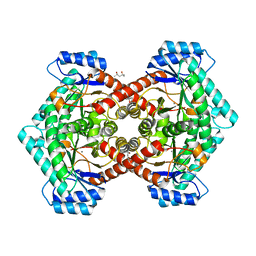 | | Structure of L-ribulose 3-epimerase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, L-ribulose 3-epimerase, MANGANESE (II) ION | | Authors: | Uechi, K, Sakuraba, H, Takata, G. | | Deposit date: | 2012-09-27 | | Release date: | 2013-10-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insight into L-ribulose 3-epimerase from Mesorhizobium loti.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2YJC
 
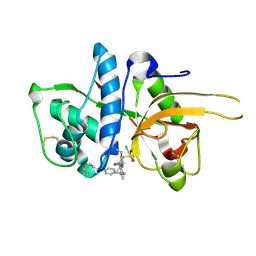 | | CATHEPSIN L WITH A NITRILE INHIBITOR | | Descriptor: | (2S,4R)-1-[1-(4-chlorophenyl)cyclopropyl]carbonyl-4-(2-chlorophenyl)sulfonyl-N-[1-(iminomethyl)cyclopropyl]pyrrolidine-2-carboxamide, CATHEPSIN L1 | | Authors: | Banner, D.W, Benz, J.M, Haap, W. | | Deposit date: | 2011-05-19 | | Release date: | 2011-11-23 | | Last modified: | 2011-11-30 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Halogen Bonding at the Active Sites of Human Cathepsin L and Mek1 Kinase: Efficient Interactions in Different Environments.
Chemmedchem, 6, 2011
|
|
2YJ9
 
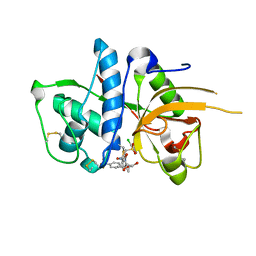 | | CATHEPSIN L WITH A NITRILE INHIBITOR | | Descriptor: | (2S,4R)-4-(2-chlorophenyl)sulfonyl-N-[1-(iminomethyl)cyclopropyl]-1-[1-[4-(trifluoromethyl)phenyl]cyclopropyl]carbonyl-pyrrolidine-2-carboxamide, CATHEPSIN L1, GLYCEROL | | Authors: | Banner, D.W, Benz, J.M, Haap, W. | | Deposit date: | 2011-05-19 | | Release date: | 2011-11-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Halogen Bonding at the Active Sites of Human Cathepsin L and Mek1 Kinase: Efficient Interactions in Different Environments.
Chemmedchem, 6, 2011
|
|
2YJ2
 
 | | CATHEPSIN L WITH A NITRILE INHIBITOR | | Descriptor: | (2S,4R)-1-[1-(4-BROMOPHENYL)CYCLOPROPYL]CARBONYL-4-(2-CHLOROPHENYL)SULFONYL-N-[1-(IMINOMETHYL)CYCLOPROPYL]PYRROLIDINE-2-CARBOXAMIDE, CATHEPSIN L1, GLYCEROL | | Authors: | Banner, D.W, Benz, J.M, Haap, W. | | Deposit date: | 2011-05-18 | | Release date: | 2011-11-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Halogen Bonding at the Active Sites of Human Cathepsin L and Mek1 Kinase: Efficient Interactions in Different Environments.
Chemmedchem, 6, 2011
|
|
2Z51
 
 | |
3WW3
 
 | | X-ray structures of Cellulomonas parahominis L-ribose isomerase with no ligand | | Descriptor: | L-ribose isomerase, MANGANESE (II) ION | | Authors: | Terami, Y, Yoshida, H, Takata, G, Kamitori, S. | | Deposit date: | 2014-06-13 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Essentiality of tetramer formation of Cellulomonas parahominis L-ribose isomerase involved in novel L-ribose metabolic pathway.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
3WW1
 
 | | X-ray structure of Cellulomonas parahominis L-ribose isomerase with L-ribose | | Descriptor: | L-ribose, L-ribose isomerase, MANGANESE (II) ION, ... | | Authors: | Terami, Y, Yoshida, H, Takata, G, Kamitori, S. | | Deposit date: | 2014-06-13 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Essentiality of tetramer formation of Cellulomonas parahominis L-ribose isomerase involved in novel L-ribose metabolic pathway.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
3WW4
 
 | | X-ray structures of Cellulomonas parahominis L-ribose isomerase with L-allose | | Descriptor: | L-allose, L-ribose isomerase, MANGANESE (II) ION, ... | | Authors: | Terami, Y, Yoshida, H, Takata, G, Kamitori, S. | | Deposit date: | 2014-06-13 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Essentiality of tetramer formation of Cellulomonas parahominis L-ribose isomerase involved in novel L-ribose metabolic pathway.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
