3ZHD
 
 | |
5EY2
 
 | | Crystal structure of CodY from Bacillus cereus | | Descriptor: | GTP-sensing transcriptional pleiotropic repressor CodY | | Authors: | Han, A, Lee, W.C, Son, J, Kim, S.H, Hwang, K.Y. | | Deposit date: | 2015-11-24 | | Release date: | 2016-09-14 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of the pleiotropic transcription regulator CodY provides insight into its GTP-sensing mechanism
Nucleic Acids Res., 44, 2016
|
|
3B23
 
 | | Crystal structure of thrombin-variegin complex: Insights of a novel mechanism of inhibition and design of tunable thrombin inhibitors | | Descriptor: | Thrombin heavy chain, Thrombin light chain, Variegin | | Authors: | Koh, C.Y, Kumar, S, Swaminathan, K, Kini, R.M. | | Deposit date: | 2011-07-20 | | Release date: | 2011-11-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of thrombin in complex with s-variegin: insights of a novel mechanism of inhibition and design of tunable thrombin inhibitors
Plos One, 6, 2011
|
|
3WKW
 
 | | Crystal structure of GH127 beta-L-arabinofuranosidase HypBA1 from Bifidobacterium longum ligand free form | | Descriptor: | Non-reducing end beta-L-arabinofuranosidase | | Authors: | Ito, T, Saikawa, K, Arakawa, T, Wakagi, T, Fujita, K. | | Deposit date: | 2013-11-01 | | Release date: | 2014-04-30 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of glycoside hydrolase family 127 beta-l-arabinofuranosidase from Bifidobacterium longum.
Biochem.Biophys.Res.Commun., 447, 2014
|
|
6MZN
 
 | |
6MZP
 
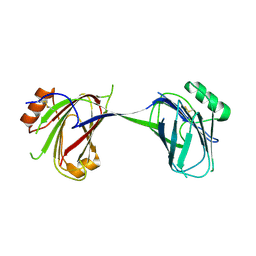 | |
3WKX
 
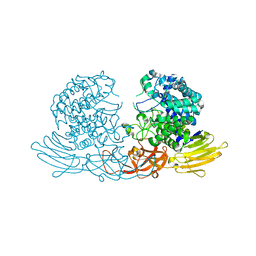 | | Crystal structure of GH127 beta-L-arabinofuranosidase HypBA1 from Bifidobacterium longum arabinose complex form | | Descriptor: | Non-reducing end beta-L-arabinofuranosidase, ZINC ION, beta-L-arabinofuranose | | Authors: | Ito, T, Saikawa, K, Arakawa, T, Wakagi, T, Fujita, K. | | Deposit date: | 2013-11-01 | | Release date: | 2014-04-30 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of glycoside hydrolase family 127 beta-l-arabinofuranosidase from Bifidobacterium longum.
Biochem.Biophys.Res.Commun., 447, 2014
|
|
4IWB
 
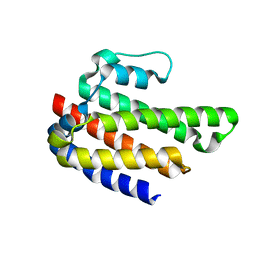 | | Novel Fold of FliC/FliS Fusion Protein | | Descriptor: | FliC, FliS chimera | | Authors: | Faham, S. | | Deposit date: | 2013-01-23 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Protein design by fusion: implications for protein structure prediction and evolution.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
8FDY
 
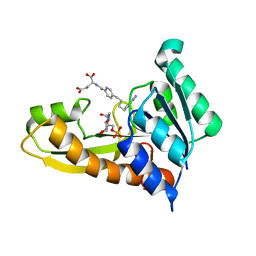 | | Human GAR transformylase in complex with GAR substrate and AGF132 inhibitor | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE, N-{5-[4-(2-amino-4-oxo-3,4-dihydrothieno[2,3-d]pyrimidin-6-yl)butyl]pyridine-2-carbonyl}-L-glutamic acid, Phosphoribosylglycinamide formyltransferase | | Authors: | Wong-Roushar, J, Dann III, C.E. | | Deposit date: | 2022-12-05 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Multitargeted 6-Substituted Thieno[2,3- d ]pyrimidines as Folate Receptor-Selective Anticancer Agents that Inhibit Cytosolic and Mitochondrial One-Carbon Metabolism.
Acs Pharmacol Transl Sci, 6, 2023
|
|
8FDX
 
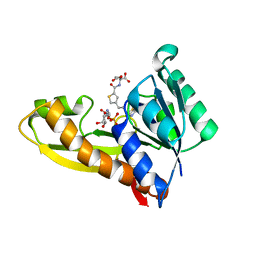 | | AGF271 and GAR in complex with human recombinant GARFTase, ligase, purine biosynthesis, transfers formyl group from 10-formyl tetrahydrofolate to glycinamide ribonucleotide (GAR) to form tetrahydrofolate and formyl GAR | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE, N-{4-[3-(2-amino-4-oxo-3,4-dihydrothieno[2,3-d]pyrimidin-6-yl)propyl]thiophene-2-carbonyl}-L-glutamic acid, Trifunctional purine biosynthetic protein adenosine-3 | | Authors: | Nyman, M.C, Wong-Roushar, J, Dann III, C.E. | | Deposit date: | 2022-12-05 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Multitargeted 6-Substituted Thieno[2,3- d ]pyrimidines as Folate Receptor-Selective Anticancer Agents that Inhibit Cytosolic and Mitochondrial One-Carbon Metabolism.
Acs Pharmacol Transl Sci, 6, 2023
|
|
8FE0
 
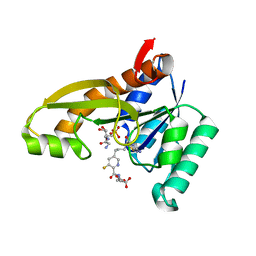 | | Human GAR transformylase in complex with GAR substrate and AGF305 inhibitor | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE, N-{5-[4-(2-amino-4-oxo-1,4-dihydrothieno[2,3-d]pyrimidin-6-yl)butyl]-3-fluoropyridine-2-carbonyl}-L-glutamic acid, Phosphoribosylglycinamide formyltransferase | | Authors: | Wong-Roushar, J, Dann III, C.E. | | Deposit date: | 2022-12-05 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Multitargeted 6-Substituted Thieno[2,3- d ]pyrimidines as Folate Receptor-Selective Anticancer Agents that Inhibit Cytosolic and Mitochondrial One-Carbon Metabolism.
Acs Pharmacol Transl Sci, 6, 2023
|
|
8FDZ
 
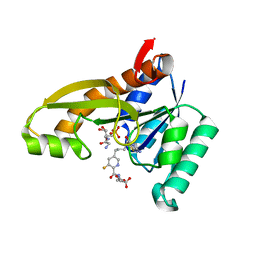 | | Human GAR transformylase in complex with GAR substrate and AGF302 inhibitor | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE, N-{5-[4-(2-amino-4-oxo-1,4-dihydrothieno[2,3-d]pyrimidin-6-yl)butyl]-3-fluoropyridine-2-carbonyl}-L-glutamic acid, Phosphoribosylglycinamide formyltransferase | | Authors: | Wong-Roushar, J, Dann III, C.E. | | Deposit date: | 2022-12-05 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Multitargeted 6-Substituted Thieno[2,3- d ]pyrimidines as Folate Receptor-Selective Anticancer Agents that Inhibit Cytosolic and Mitochondrial One-Carbon Metabolism.
Acs Pharmacol Transl Sci, 6, 2023
|
|
6AIY
 
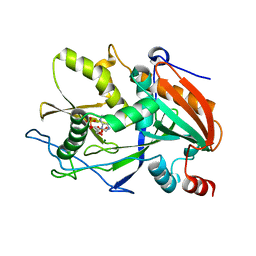 | |
6AIX
 
 | |
7DCH
 
 | | Alpha-glucosidase from Weissella cibaria BBK-1 bound with acarbose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-glycosidase, ... | | Authors: | Krusong, K, Wangpaiboon, K, Kim, S, Mori, T, Hakoshima, T. | | Deposit date: | 2020-10-26 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.692 Å) | | Cite: | A GH13 alpha-glucosidase from Weissella cibaria uncommonly acts on short-chain maltooligosaccharides.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7D9C
 
 | | Alpha-glucosidase from Weissella cibaria BBK-1 bound with maltose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Alpha-glycosidase, CALCIUM ION, ... | | Authors: | Krusong, K, Wangpaiboon, K, Kim, S, Mori, T, Hakoshima, T. | | Deposit date: | 2020-10-13 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | A GH13 alpha-glucosidase from Weissella cibaria uncommonly acts on short-chain maltooligosaccharides.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7D9B
 
 | | Crystal structure of alpha-glucosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Alpha-glycosidase, CALCIUM ION, ... | | Authors: | Krusong, K, Wangpaiboon, K, Kim, S, Mori, T, Hakoshima, T. | | Deposit date: | 2020-10-12 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | A GH13 alpha-glucosidase from Weissella cibaria uncommonly acts on short-chain maltooligosaccharides.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7DCG
 
 | | Alpha-glucosidase from Weissella cibaria BBK-1 bound with maltotriose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Alpha-glycosidase, CALCIUM ION, ... | | Authors: | Krusong, K, Wangpaiboon, K, Kim, S, Mori, T, Hakoshima, T. | | Deposit date: | 2020-10-26 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | A GH13 alpha-glucosidase from Weissella cibaria uncommonly acts on short-chain maltooligosaccharides.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6BNR
 
 | | Carbonmonoxy hemoglobin in complex with the antisickling agent 5-methoxy-2-(pyridin-2-ylmethoxy)benzaldehyde (INN310) | | Descriptor: | 2-[(4-methoxy-2-methylphenoxy)methyl]pyridine, CARBON MONOXIDE, Hemoglobin subunit alpha, ... | | Authors: | Pagare, P.P, Musayev, F.N, Safo, M.K. | | Deposit date: | 2017-11-17 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Rational design of pyridyl derivatives of vanillin for the treatment of sickle cell disease.
Bioorg. Med. Chem., 26, 2018
|
|
4O99
 
 | | Crystal structure of Beta-ketothiolase (PhaA) from Ralstonia eutropha H16 | | Descriptor: | Acetyl-CoA acetyltransferase, GLYCEROL | | Authors: | Kim, E.J, Kim, J, Kim, S, Kim, K.J. | | Deposit date: | 2014-01-02 | | Release date: | 2014-12-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure and biochemical characterization of PhaA from Ralstonia eutropha, a polyhydroxyalkanoate-producing bacterium.
Biochem.Biophys.Res.Commun., 452, 2014
|
|
4O9C
 
 | | Crystal structure of Beta-ketothiolase (PhaA) from Ralstonia eutropha H16 | | Descriptor: | Acetyl-CoA acetyltransferase, COENZYME A | | Authors: | Kim, E.J, Kim, J, Kim, S, Kim, K.J. | | Deposit date: | 2014-01-02 | | Release date: | 2014-12-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and biochemical characterization of PhaA from Ralstonia eutropha, a polyhydroxyalkanoate-producing bacterium.
Biochem.Biophys.Res.Commun., 452, 2014
|
|
1OIL
 
 | | STRUCTURE OF LIPASE | | Descriptor: | CALCIUM ION, LIPASE | | Authors: | Kim, K.K, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 1996-12-06 | | Release date: | 1997-05-15 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of a triacylglycerol lipase from Pseudomonas cepacia reveals a highly open conformation in the absence of a bound inhibitor.
Structure, 5, 1997
|
|
4O9A
 
 | | Crystal structure of Beta-ketothiolase (PhaA) from Ralstonia eutropha H16 | | Descriptor: | Acetyl-CoA acetyltransferase | | Authors: | Kim, E.J, Kim, J, Kim, S, Kim, K.J. | | Deposit date: | 2014-01-02 | | Release date: | 2015-02-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal structure and biochemical characterization of PhaA from Ralstonia eutropha, a polyhydroxyalkanoate-producing bacterium.
Biochem.Biophys.Res.Commun., 452, 2014
|
|
1QZY
 
 | | Human Methionine Aminopeptidase in complex with bengamide inhibitor LAF153 and cobalt | | Descriptor: | (E)-(2R,3R,4S,5R)-3,4,5-TRIHYDROXY-2-METHOXY-8,8-DIMETHYL-NON-6-ENOIC ACID ((3S,6R)-6-HYDROXY-2-OXO-AZEPAN-3-YL)-AMIDE, COBALT (II) ION, Methionine aminopeptidase 2, ... | | Authors: | Eck, M.J, Song, H.K, Morollo, A. | | Deposit date: | 2003-09-18 | | Release date: | 2003-11-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Proteomics-based target identification: bengamides as a new class of methionine aminopeptidase inhibitors.
J.Biol.Chem., 278, 2003
|
|
7EHI
 
 | | Crystal structure of covalent maltosyl-alpha-glucosidase intermediate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Krusong, K, Wangpaiboon, K, Kim, S, Mori, T, Hakoshima, T. | | Deposit date: | 2021-03-29 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A GH13 alpha-glucosidase from Weissella cibaria uncommonly acts on short-chain maltooligosaccharides.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
