6U8Q
 
 | | CryoEM structure of HIV-1 cleaved synaptic complex (CSC) intasome | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Li, M, Chen, X, Craigie, R. | | Deposit date: | 2019-09-05 | | Release date: | 2020-02-05 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (4.67 Å) | | Cite: | A Peptide Derived from Lens Epithelium-Derived Growth Factor Stimulates HIV-1 DNA Integration and Facilitates Intasome Structural Studies.
J.Mol.Biol., 432, 2020
|
|
6VDK
 
 | | CryoEM structure of HIV-1 conserved Intasome Core | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Li, M, Chen, X, Craigie, R. | | Deposit date: | 2019-12-27 | | Release date: | 2020-02-05 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | A Peptide Derived from Lens Epithelium-Derived Growth Factor Stimulates HIV-1 DNA Integration and Facilitates Intasome Structural Studies.
J.Mol.Biol., 432, 2020
|
|
7F16
 
 | | Cryo-EM structure of parathyroid hormone receptor type 2 in complex with a tuberoinfundibular peptide of 39 residues and G protein | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, X, Cheng, X, Zhao, L.H, Wang, Y, Ye, C, Zou, X, Dai, A, Cong, Z, Chen, J, Zhou, Q, Xia, T, Jiang, H, Xu, H.E, Yang, D, Wang, M.W. | | Deposit date: | 2021-06-08 | | Release date: | 2021-08-18 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular insights into differentiated ligand recognition of the human parathyroid hormone receptor 2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
9IV9
 
 | | Cryo-EM structure of a truncated Nipah Virus L Protein bound by Phosphoprotein Tetramer | | Descriptor: | Phosphoprotein, RNA-directed RNA polymerase L, ZINC ION | | Authors: | Xue, L, Chang, T, Gui, J, Li, Z, Zhao, H, Zou, B, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-07-23 | | Release date: | 2025-05-21 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.31 Å) | | Cite: | Cryo-EM structures of Nipah virus polymerase complex reveal highly varied interactions between L and P proteins among paramyxoviruses.
Protein Cell, 2025
|
|
9IVA
 
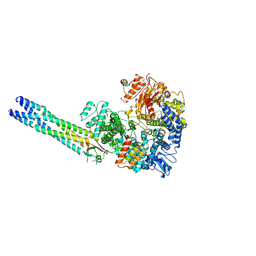 | | Cryo-EM structure of the full-length Nipah Virus L Protein bound by Phosphoprotein Tetramer | | Descriptor: | Phosphoprotein, RNA-directed RNA polymerase L, ZINC ION | | Authors: | Xue, L, Chang, T, Gui, J, Li, Z, Zhao, H, Zou, B, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-07-23 | | Release date: | 2025-05-21 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Cryo-EM structures of Nipah virus polymerase complex reveal highly varied interactions between L and P proteins among paramyxoviruses.
Protein Cell, 2025
|
|
2JQQ
 
 | | Solution structure of Saccharomyces cerevisiae conserved oligomeric Golgi subunit 2 protein (Cog2p) | | Descriptor: | Conserved oligomeric Golgi complex subunit 2 | | Authors: | Cavanaugh, L.F, Chen, X, Pelczer, I, Rizo, J, Hughson, F.M. | | Deposit date: | 2007-06-06 | | Release date: | 2007-06-19 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of conserved oligomeric Golgi complex subunit 2
J.Biol.Chem., 282, 2007
|
|
9JVP
 
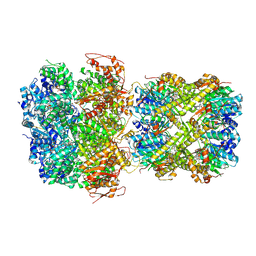 | | CryoEM structure of M. tuberculosis ClpC1P1P2 complex bound to bortezomib, conformation 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpC1, ... | | Authors: | Zhou, B, Zhao, H, Gao, Y, Chen, X, Zhang, T, He, J, Xiong, X. | | Deposit date: | 2024-10-09 | | Release date: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.15 Å) | | Cite: | Activation mechanism of caseinolytic chaperone-protease system in Mycobacterium tuberculosis by the anti-cancer drug bortezomib
To Be Published
|
|
2KVT
 
 | | solution NMR structure of yaiA from Escherichia Eoli. Northeast Structural Genomics Target ER244 | | Descriptor: | Uncharacterized protein yaiA | | Authors: | Tang, Y, Chen, X, Ciccosanti, C, Janjua, H, Xiao, R, Acton, T, Everett, J, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-03-28 | | Release date: | 2010-05-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | solution NMR structure of yaiA from Escherichia coli. Northeast Structural Genomics Target ER244
To be Published
|
|
9BKX
 
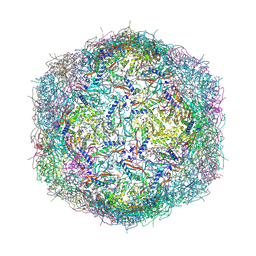 | | Mycobacterium tuberculosis encapsulin in complex with DyP | | Descriptor: | DODECAETHYLENE GLYCOL, Dye-decolorizing peroxidase, GLYCEROL, ... | | Authors: | Cuthbert, B.J, Batot, G.O, Contreras, H, Chen, X, Burley, K.H, Goulding, C.W. | | Deposit date: | 2024-04-29 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural Characterization of Mycobacterium tuberculosis Encapsulin in Complex with Dye-Decolorizing Peroxide.
Microorganisms, 12, 2024
|
|
3V78
 
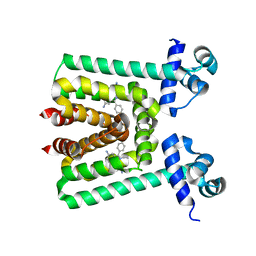 | | Crystal Structure of Transcriptional Regulator | | Descriptor: | ETHIDIUM, PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN (PROBABLY DEOR-FAMILY) | | Authors: | Do, S.V, Bolla, J.R, Chen, X, Yu, E.W. | | Deposit date: | 2011-12-20 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Structural and functional analysis of the transcriptional regulator Rv3066 of Mycobacterium tuberculosis.
Nucleic Acids Res., 40, 2012
|
|
3T6N
 
 | |
3V6G
 
 | | Crystal Structure of Transcriptional Regulator | | Descriptor: | PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN (PROBABLY DEOR-FAMILY) | | Authors: | Do, S.V, Bolla, J.R, Chen, X, Yu, E.W. | | Deposit date: | 2011-12-19 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.821 Å) | | Cite: | Structural and functional analysis of the transcriptional regulator Rv3066 of Mycobacterium tuberculosis.
Nucleic Acids Res., 40, 2012
|
|
4GHT
 
 | | Crystal structure of EV71 3C proteinase in complex with AG7088 | | Descriptor: | 3C proteinase, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | Authors: | Chen, C, Wu, C, Cai, Q, Li, N, Peng, X, Cai, Y, Yin, K, Chen, X, Wang, X, Zhang, R, Liu, L, Chen, S, Li, J, Lin, T. | | Deposit date: | 2012-08-08 | | Release date: | 2013-06-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structures of Enterovirus 71 3C proteinase (strain E2004104-TW-CDC) and its complex with rupintrivir
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GHQ
 
 | | Crystal structure of EV71 3C proteinase | | Descriptor: | 3C proteinase | | Authors: | Chen, C, Wu, C, Cai, Q, Li, N, Peng, X, Cai, Y, Yin, K, Chen, X, Wang, X, Zhang, R, Liu, L, Chen, S, Li, J, Lin, T. | | Deposit date: | 2012-08-08 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of Enterovirus 71 3C proteinase (strain E2004104-TW-CDC) and its complex with rupintrivir
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4NKJ
 
 | |
1U2Z
 
 | | Crystal structure of histone K79 methyltransferase Dot1p from yeast | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Sawada, K, Yang, Z, Horton, J.R, Collins, R.E, Zhang, X, Cheng, X. | | Deposit date: | 2004-07-20 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the conserved core of the yeast Dot1p, a nucleosomal histone H3 lysine 79 methyltransferase
J.Biol.Chem., 279, 2004
|
|
7DL2
 
 | | Cryo-EM structure of human TSC complex | | Descriptor: | Hamartin, Isoform 7 of Tuberin, TBC1 domain family member 7, ... | | Authors: | Yang, H, Yu, Z, Chen, X, Li, J, Li, N, Cheng, J, Gao, N, Yuan, H, Ye, D, Guan, K, Xu, Y. | | Deposit date: | 2020-11-25 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural insights into TSC complex assembly and GAP activity on Rheb.
Nat Commun, 12, 2021
|
|
7YE9
 
 | | SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-05 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
4ICK
 
 | | Crystal structure of human AP4A hydrolase E58A mutant | | Descriptor: | Bis(5'-nucleosyl)-tetraphosphatase [asymmetrical], GLYCEROL, MAGNESIUM ION, ... | | Authors: | Ge, H, Chen, X. | | Deposit date: | 2012-12-10 | | Release date: | 2013-12-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of wild-type and mutant human Ap4A hydrolase.
Biochem.Biophys.Res.Commun., 432, 2013
|
|
7YDY
 
 | | SARS-CoV-2 Spike (6P) in complex with 1 R1-32 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-04 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.75 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
4IJX
 
 | |
7YEG
 
 | | SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs and 3 ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-05 | | Release date: | 2022-08-24 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
7YDI
 
 | | SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs and 3 ACE2, focused refinement of RBD region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32, Light chain of R1-32, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-04 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
4R36
 
 | | Crystal structure analysis of LpxA, a UDP-N-acetylglucosamine acyltransferase from Bacteroides fragilis 9343 | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ACETATE ION, ... | | Authors: | Ngo, A, Fong, K, Cox, D, Fisher, A, Chen, X. | | Deposit date: | 2014-08-14 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Bacteroides fragilis uridine 5'-diphosphate-N-acetylglucosamine (UDP-GlcNAc) acyltransferase (BfLpxA).
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7YE5
 
 | | SARS-CoV-2 Spike (6P) in complex with 2 R1-32 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-05 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (6.75 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
