5YQN
 
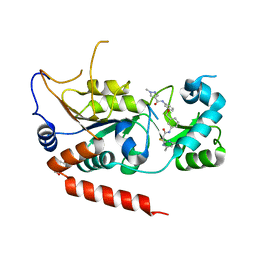 | | Crystal structure of Sirt2 in complex with selective inhibitor L55 | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-[3-[[3-[2-(4,6-dimethylpyrimidin-2-yl)sulfanylethanoylamino]phenyl]methoxy]phenyl]-1-methyl-pyrazole-4-carboxamide, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Wang, H, Yu, Y, Li, G, Chen, Q. | | Deposit date: | 2017-11-07 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray crystal structure guided discovery of new selective, substrate-mimicking sirtuin 2 inhibitors that exhibit activities against non-small cell lung cancer cells.
Eur J Med Chem, 155, 2018
|
|
5YQO
 
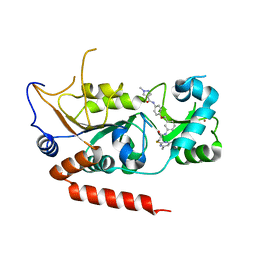 | | Crystal structure of Sirt2 in complex with selective inhibitor L5C | | Descriptor: | N-[4-[[3-[2-(4,6-dimethylpyrimidin-2-yl)sulfanylethanoylamino]phenyl]methoxy]phenyl]-1-methyl-pyrazole-4-carboxamide, NAD-dependent protein deacetylase sirtuin-2, ZINC ION | | Authors: | Wang, H, Yu, Y, Li, G, Chen, Q. | | Deposit date: | 2017-11-07 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.483 Å) | | Cite: | X-ray crystal structure guided discovery of new selective, substrate-mimicking sirtuin 2 inhibitors that exhibit activities against non-small cell lung cancer cells.
Eur J Med Chem, 155, 2018
|
|
5YQL
 
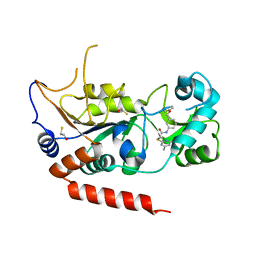 | | Crystal structure of Sirt2 in complex with selective inhibitor A2I | | Descriptor: | 2-(4,6-dimethylpyrimidin-2-yl)sulfanyl-N-[3-(phenoxymethyl)phenyl]ethanamide, BETA-MERCAPTOETHANOL, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Wang, H, Yu, Y, Li, G, Chen, Q. | | Deposit date: | 2017-11-07 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | X-ray crystal structure guided discovery of new selective, substrate-mimicking sirtuin 2 inhibitors that exhibit activities against non-small cell lung cancer cells.
Eur J Med Chem, 155, 2018
|
|
5YQM
 
 | | Crystal structure of Sirt2 in complex with selective inhibitor A29 | | Descriptor: | 2-(4,6-dimethylpyrimidin-2-yl)sulfanyl-N-(4-phenylsulfanylphenyl)ethanamide, BETA-MERCAPTOETHANOL, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Wang, H, Yu, Y, Li, G, chen, Q. | | Deposit date: | 2017-11-07 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.735 Å) | | Cite: | X-ray crystal structure guided discovery of new selective, substrate-mimicking sirtuin 2 inhibitors that exhibit activities against non-small cell lung cancer cells.
Eur J Med Chem, 155, 2018
|
|
7VAH
 
 | |
5NSE
 
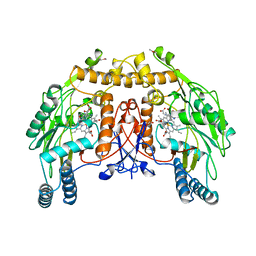 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE, H4B-FREE, HYDROXY-ARG COMPLEX | | Descriptor: | ACETATE ION, CACODYLIC ACID, GLYCEROL, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 1999-01-13 | | Release date: | 2002-05-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of the Heme Domain of Bovine Endothelial Nitric Oxide Synthase Complexed with Arginine Analogues
To be Published
|
|
6NSE
 
 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE, H4B-FREE, CANAVANINE COMPLEX | | Descriptor: | ACETATE ION, CACODYLIC ACID, GLYCEROL, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 1999-01-13 | | Release date: | 2002-05-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structures of the Heme Domain of Bovine Endothelial Nitric Oxide Synthase Complexed with Arginine Analogues
To be Published
|
|
4X2R
 
 | | Crystal structure of PriA from Actinomyces urogenitalis | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, PHOSPHATE ION | | Authors: | MICHALSKA, K, VERDUZCO-CASTRO, E.A, ENDRES, M, BARONA-GOMEZ, F, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-11-26 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Evolution of substrate specificity in a retained enzyme driven by gene loss.
Elife, 6, 2017
|
|
4AGW
 
 | | Discovery of a small molecule type II inhibitor of wild-type and gatekeeper mutants of BCR-ABL, PDGFRalpha, Kit, and Src kinases | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-{2-[(cyclopropylcarbonyl)amino][1,3]thiazolo[5,4-b]pyridin-5-yl}-N-{4-[(4-ethylpiperazin-1-yl)methyl]-3-(trifluoromet hyl)phenyl}benzamide, GLYCEROL, ... | | Authors: | Weisberg, E, Choi, H.G, Seeliger, M, Gray, N, Griffin, J.D. | | Deposit date: | 2012-02-01 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of a Small-Molecule Type II Inhibitor of Wild-Type and Gatekeeper Mutants of Bcr-Abl, Pdgfralpha, Kit, and Src Kinases: Novel Type II Inhibitor of Gatekeeper Mutants.
Blood, 115, 2010
|
|
7WV9
 
 | | Allosteric modulator ZCZ011 binding to CP55940-bound cannabinoid receptor 1 in complex with Gi protein | | Descriptor: | 2-[(1R,2R,5R)-5-hydroxy-2-(3-hydroxypropyl)cyclohexyl]-5-(2-methyloctan-2-yl)phenol, 6-methyl-3-[(1S)-2-nitro-1-thiophen-2-yl-ethyl]-2-phenyl-1H-indole, Cannabinoid receptor 1, ... | | Authors: | Xu, Z, Shao, Z. | | Deposit date: | 2022-02-10 | | Release date: | 2022-06-15 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Molecular mechanism of allosteric modulation for the cannabinoid receptor CB1.
Nat.Chem.Biol., 18, 2022
|
|
7C4U
 
 | | MicroED structure of orthorhombic Vancomycin at 1.2 A resolution | | Descriptor: | CHLORIDE ION, Vancomycin, vancosamine-(1-2)-beta-D-glucopyranose | | Authors: | Fan, Q, Zhou, H, Li, X, Wang, J. | | Deposit date: | 2020-05-18 | | Release date: | 2020-08-12 | | Last modified: | 2025-04-09 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.2 Å) | | Cite: | Precise Control Over Kinetics of Molecular Assembly: Production of Particles with Tunable Sizes and Crystalline Forms.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7C4V
 
 | | MicroED structure of anorthic Vancomycin at 1.05 A resolution | | Descriptor: | CHLORIDE ION, Vancomycin, vancosamine-(1-2)-beta-D-glucopyranose | | Authors: | Fan, Q, Zhou, H, Li, X, Wang, J. | | Deposit date: | 2020-05-18 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-29 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.05 Å) | | Cite: | Precise Control Over Kinetics of Molecular Assembly: Production of Particles with Tunable Sizes and Crystalline Forms.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
3KMU
 
 | |
1NSI
 
 | | HUMAN INDUCIBLE NITRIC OXIDE SYNTHASE, ZN-BOUND, L-ARG COMPLEX | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ARGININE, GLYCEROL, ... | | Authors: | Li, H, Raman, C.S, Glaser, C.B, Blasko, E, Young, T.A, Parkinson, J.F, Whitlow, M, Poulos, T.L. | | Deposit date: | 1999-01-10 | | Release date: | 2000-01-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structures of zinc-free and -bound heme domain of human inducible nitric-oxide synthase. Implications for dimer stability and comparison with endothelial nitric-oxide synthase.
J.Biol.Chem., 274, 1999
|
|
1CLK
 
 | | CRYSTAL STRUCTURE OF STREPTOMYCES DIASTATICUS NO.7 STRAIN M1033 XYLOSE ISOMERASE AT 1.9 A RESOLUTION WITH PSEUDO-I222 SPACE GROUP | | Descriptor: | COBALT (II) ION, MAGNESIUM ION, XYLOSE ISOMERASE | | Authors: | Niu, L, Teng, M, Zhu, X, Gong, W. | | Deposit date: | 1999-04-29 | | Release date: | 2000-05-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of xylose isomerase from Streptomyces diastaticus no. 7 strain M1033 at 1.85 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
7CRH
 
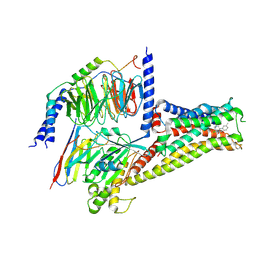 | | Cryo-EM structure of SKF83959 bound dopamine receptor DRD1-Gs signaling complex | | Descriptor: | (1S)-6-chloranyl-3-methyl-1-(3-methylphenyl)-1,2,4,5-tetrahydro-3-benzazepine-7,8-diol, D(1A) dopamine receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Yan, W, Shao, Z.H. | | Deposit date: | 2020-08-13 | | Release date: | 2021-03-03 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Ligand recognition and allosteric regulation of DRD1-Gs signaling complexes.
Cell, 184, 2021
|
|
7CKW
 
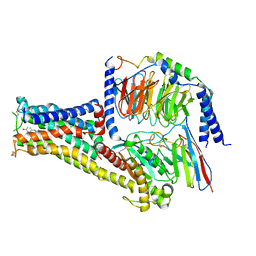 | | Cryo-EM structure of Fenoldopam bound dopamine receptor DRD1-Gs signaling complex | | Descriptor: | (1R)-6-chloranyl-1-(4-hydroxyphenyl)-2,3,4,5-tetrahydro-1H-3-benzazepine-7,8-diol, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Yan, W, Shao, W. | | Deposit date: | 2020-07-20 | | Release date: | 2021-03-03 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Ligand recognition and allosteric regulation of DRD1-Gs signaling complexes.
Cell, 184, 2021
|
|
7CKY
 
 | | Cryo-EM structure of PW0464 bound dopamine receptor DRD1-Gs signaling complex | | Descriptor: | 6-[4-[3-[bis(fluoranyl)methoxy]pyridin-2-yl]oxy-2-methyl-phenyl]-1,5-dimethyl-pyrimidine-2,4-dione, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Yan, W, Shao, Z. | | Deposit date: | 2020-07-20 | | Release date: | 2021-03-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Ligand recognition and allosteric regulation of DRD1-Gs signaling complexes.
Cell, 184, 2021
|
|
7CKZ
 
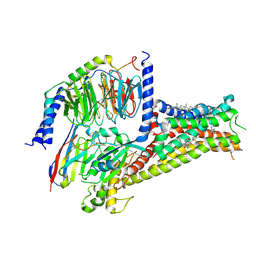 | | Cryo-EM structure of Dopamine and LY3154207 bound dopamine receptor DRD1-Gs signaling complex | | Descriptor: | 2-[2,6-bis(chloranyl)phenyl]-1-[(1S,3R)-3-(hydroxymethyl)-1-methyl-5-(3-methyl-3-oxidanyl-butyl)-3,4-dihydro-1H-isoquinolin-2-yl]ethanone, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Yan, W, Shao, Z. | | Deposit date: | 2020-07-20 | | Release date: | 2021-03-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Ligand recognition and allosteric regulation of DRD1-Gs signaling complexes.
Cell, 184, 2021
|
|
7CKX
 
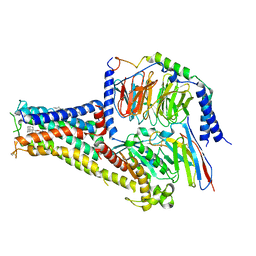 | | Cryo-EM structure of A77636 bound dopamine receptor DRD1-Gs signaling complex | | Descriptor: | (1R,3S)-3-(1-adamantyl)-1-(aminomethyl)-3,4-dihydro-1H-isochromene-5,6-diol, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Yan, W, Shao, Z. | | Deposit date: | 2020-07-20 | | Release date: | 2021-03-03 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Ligand recognition and allosteric regulation of DRD1-Gs signaling complexes.
Cell, 184, 2021
|
|
3KMW
 
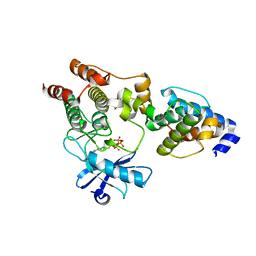 | | Crystal structure of the ILK/alpha-parvin core complex (MgATP) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Alpha-parvin, Integrin-linked kinase, ... | | Authors: | Fukuda, K, Qin, J. | | Deposit date: | 2009-11-11 | | Release date: | 2009-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The pseudoactive site of ILK is essential for its binding to alpha-Parvin and localization to focal adhesions.
Mol.Cell, 36, 2009
|
|
7DVP
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp4|5 peptidyl substrate | | Descriptor: | 3C-like proteinase, nsp4/5 peptidyl substrate | | Authors: | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | Deposit date: | 2021-01-14 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DW0
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp14|15 peptidyl substrate | | Descriptor: | 3C-like proteinase, nsp14/15 peptidyl substrate | | Authors: | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | Deposit date: | 2021-01-15 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DW6
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp15|16 peptidyl substrate | | Descriptor: | 3C-like proteinase, nsp15/16 peptidyl substrate | | Authors: | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | Deposit date: | 2021-01-15 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DVY
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp9|10 peptidyl substrate | | Descriptor: | 3C-like proteinase, nsp9/10 peptidyl substrate | | Authors: | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | Deposit date: | 2021-01-15 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
