3USD
 
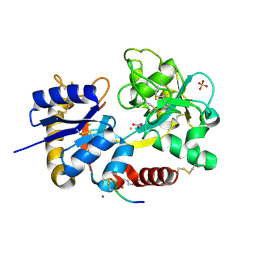 | | Crystal Structure of C-lobe of Bovine lactoferrin Complexed with Imidazol (1,2 a) pyridine3-yl-acitic acid at 2.4 A Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C-terminal peptide of Lactotransferrin, ... | | Authors: | Shukla, P.K, Gautam, L, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-11-23 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of C-lobe of Bovine lactoferrin Complexed with Imidazol (1,2 a) pyridine3-yl-acitic acid at 2.4 A Resolution
To be Published
|
|
3V05
 
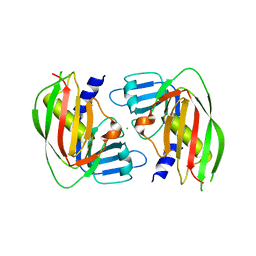 | | 2.4 Angstrom Crystal Structure of Superantigen-like Protein from Staphylococcus aureus. | | Descriptor: | CHLORIDE ION, Superantigen-like Protein | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Filippova, E.V, Dubrovska, I, Winsor, J, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-12-07 | | Release date: | 2011-12-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2.4 Angstrom Crystal Structure of Superantigen-like Protein from Staphylococcus aureus.
TO BE PUBLISHED
|
|
3UU4
 
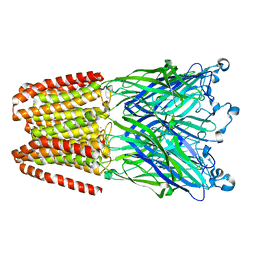 | | The GLIC pentameric Ligand-Gated Ion Channel Loop2-21' mutant reduced in the crystal in a locally-closed conformation (LC1 subtype) | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Glr4197 protein | | Authors: | Sauguet, L, Nury, H, Corringer, P.J, Delarue, M. | | Deposit date: | 2011-11-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | A locally closed conformation of a bacterial pentameric proton-gated ion channel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3V0D
 
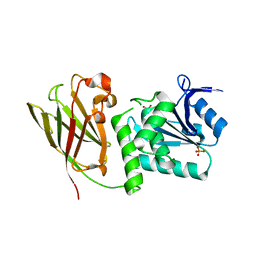 | | Crystal structure of Ciona intestinalis voltage sensor-containing phosphatase (Ci-VSP), residues 241-576(C363S) | | Descriptor: | PHOSPHATE ION, Voltage-sensor containing phosphatase | | Authors: | Liu, L, Kohout, S.C, Xu, Q, Muller, S, Kimberlin, C, Isacoff, E.Y, Minor, D.L. | | Deposit date: | 2011-12-07 | | Release date: | 2012-05-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | A glutamate switch controls voltage-sensitive phosphatase function.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3V2U
 
 | | Crystal structure of the yeast GAL regulon complex of the repressor, Gal80p, and the transducer, Gal3p, with galactose and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, Galactose/lactose metabolism regulatory protein GAL80, ... | | Authors: | Lavy, T, Kumar, P.R, He, H, Joshua-Tor, L. | | Deposit date: | 2011-12-12 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | The Gal3p transducer of the GAL regulon interacts with the Gal80p repressor in its ligand-induced closed conformation.
Genes Dev., 26, 2012
|
|
3V70
 
 | | Crystal Structure of Human GTPase IMAP family member 1 | | Descriptor: | GTPase IMAP family member 1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Nedyalkova, L, Shen, Y, Tong, Y, Tempel, W, Mackenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Andrews, D.W, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-12-20 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Crystal Structure of Human GTPase IMAP family member 1
to be published
|
|
3UID
 
 | | Crystal Structure of Protein Ms6760 from Mycobacterium smegmatis | | Descriptor: | Putative uncharacterized protein | | Authors: | Bajaj, R.A, Miallau, L, Cascio, D, Arbing, M, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2011-11-04 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.571 Å) | | Cite: | Crystal structure of the toxin Msmeg_6760, the structural homolog of Mycobacterium tuberculosis Rv2035, a novel type II toxin involved in the hypoxic response.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
6KPF
 
 | | Cryo-EM structure of a class A GPCR with G protein complex | | Descriptor: | 7-[(6aR,9R,10aR)-1-Hydroxy-9-(hydroxymethyl)-6,6-dimethyl-6a,7,8,9,10,10a-hexahydro-6H-benzo[c]chromen-3-yl]- 7-methyloctanenitrile, Cannabinoid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Li, X.T, Hua, T, Wu, L.J, Makriyannis, A, Shen, L, Wang, Y.X, Liu, Z.J. | | Deposit date: | 2019-08-15 | | Release date: | 2020-02-12 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Activation and Signaling Mechanism Revealed by Cannabinoid Receptor-GiComplex Structures.
Cell, 180, 2020
|
|
3V4A
 
 | | Structure of ar lbd with activator peptide and sarm inhibitor 2 | | Descriptor: | (5R)-3-(3,4-dichlorophenyl)-5-(4-hydroxyphenyl)-1,5-dimethyl-2-thioxoimidazolidin-4-one, Androgen receptor, SULFATE ION | | Authors: | Nique, F, Hebbe, S, Peixoto, C, Annoot, D, Lefrancois, J.-M, Duval, E, Michoux, L, Triballeau, N, Lemoullec, J.M, Mollat, P, Thauvin, M, Prange, T, Minet, D, Clement-Lacroix, P, Robin-Jagerschmidt, C, Fleury, D, Guedin, D, Deprez, P. | | Deposit date: | 2011-12-14 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of diarylhydantoins as new selective androgen receptor modulators.
J.Med.Chem., 55, 2012
|
|
3V36
 
 | | Aldose reductase complexed with glceraldehyde | | Descriptor: | Aldose reductase, D-Glyceraldehyde, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Zheng, X, Zhang, L, Chen, Y, Luo, H, Hu, X. | | Deposit date: | 2011-12-13 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Partial inhibition of aldose reductase by nitazoxanide and its molecular basis.
Chemmedchem, 7, 2012
|
|
3V4Z
 
 | | D-alanine--D-alanine ligase from Yersinia pestis | | Descriptor: | D-alanine--D-alanine ligase, DI(HYDROXYETHYL)ETHER, TRIETHYLENE GLYCOL | | Authors: | Osipiuk, J, Nocek, B, Mulligan, R, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-12-15 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | D-alanine--D-alanine ligase from Yersinia pestis.
To be Published
|
|
7R6Z
 
 | | OXA-48 bound by Compound 3.3 | | Descriptor: | 1,2-ETHANEDIOL, 4-amino-5-hydroxynaphthalene-2,7-disulfonic acid, Beta-lactamase, ... | | Authors: | Taylor, D.M, Hu, L, Prasad, B.V.V, Sankaran, B, Palzkill, T. | | Deposit date: | 2021-06-24 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unique Diacidic Fragments Inhibit the OXA-48 Carbapenemase and Enhance the Killing of Escherichia coli Producing OXA-48.
Acs Infect Dis., 7, 2021
|
|
6KPG
 
 | | Cryo-EM structure of CB1-G protein complex | | Descriptor: | (6~{a}~{R},9~{R},10~{a}~{R})-9-(hydroxymethyl)-3-(8-isothiocyanato-2-methyl-octan-2-yl)-6,6-dimethyl-6~{a},7,8,9,10,10~{a}-hexahydrobenzo[c]chromen-1-ol, Cannabinoid receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Hua, T, Li, X.T, Wu, L.J, Makriyannis, A, Wang, Y.X, Shen, L, Liu, Z.J. | | Deposit date: | 2019-08-15 | | Release date: | 2020-02-12 | | Last modified: | 2020-03-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Activation and Signaling Mechanism Revealed by Cannabinoid Receptor-GiComplex Structures.
Cell, 180, 2020
|
|
6KVM
 
 | | Crystal structure of Chicken MHC Class II for 1.9 angstrom | | Descriptor: | GLYCEROL, MHC class II alpha chain, MHC class II beta chain 2, ... | | Authors: | Zhang, L, Xia, C. | | Deposit date: | 2019-09-04 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | A Newly Recognized Pairing Mechanism of the alpha- and beta-Chains of the Chicken Peptide-MHC Class II Complex.
J Immunol., 204, 2020
|
|
6KYB
 
 | | Crystal structure of Atg18 from Saccharomyces cerevisiae | | Descriptor: | Autophagy-related protein 18 | | Authors: | Tang, D, Lei, Y, Liao, G, Chen, Q, Xu, L, Lu, K, Qi, S. | | Deposit date: | 2019-09-17 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of Atg18 reveals a new binding site for Atg2 in Saccharomyces cerevisiae.
Cell.Mol.Life Sci., 78, 2021
|
|
7XYO
 
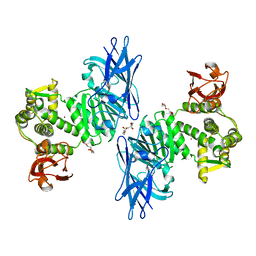 | | Crystal Structure of a M61 aminopeptidase family member from Myxococcus fulvus | | Descriptor: | Aminopeptidase M61, HEXAETHYLENE GLYCOL, TETRAETHYLENE GLYCOL, ... | | Authors: | Chen, X, Wang, X, Huo, L, Wu, D. | | Deposit date: | 2022-06-02 | | Release date: | 2023-06-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery and Characterization of a Myxobacterial Lanthipeptide with Unique Biosynthetic Features and Anti-inflammatory Activity.
J.Am.Chem.Soc., 145, 2023
|
|
2XN1
 
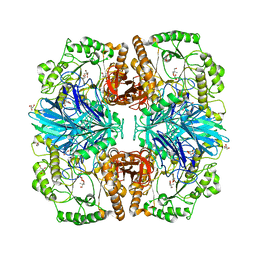 | | Structure of alpha-galactosidase from Lactobacillus acidophilus NCFM with TRIS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ALPHA-GALACTOSIDASE, GLYCEROL | | Authors: | Fredslund, F, Abou Hachem, M, Larsen, R.J, Sorensen, P.G, Lo Leggio, L, Svensson, B. | | Deposit date: | 2010-07-30 | | Release date: | 2011-08-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Alpha-Galactosidase from Lactobacillus Acidophilus Ncfm: Insight Into Tetramer Formation and Substrate Binding.
J.Mol.Biol., 412, 2011
|
|
8ZAV
 
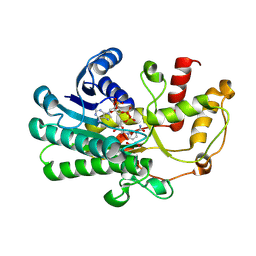 | | alcohol dehydrogenases KpADH mutant - S9Y/F161K | | Descriptor: | 1,2-ETHANEDIOL, NAD-dependent epimerase/dehydratase domain-containing protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, L, Ni, Y, Xu, G.C. | | Deposit date: | 2024-04-25 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Engineering alcohol dehydrogenases KpADH for enhanced organic-solvent tolerance and its molecular mechanisms
To Be Published
|
|
8Z85
 
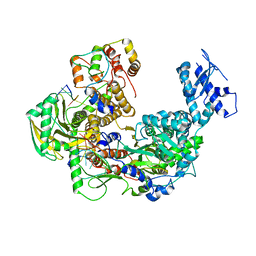 | | Cryo-EM structure of Thogoto virus polymerase in transcription pre-initiation conformation 1 | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-R(*AP*GP*AP*GP*AP*AP*AP*UP*CP*AP*AP*GP*GP*CP*AP*GP*UP*U)-3'), ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-21 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
9BBI
 
 | | Co-crystal structure of human DDB1 bound to fragment UB028669 | | Descriptor: | 3-([1,3]oxazolo[4,5-b]pyridin-2-yl)aniline, DNA damage-binding protein 1, L(+)-TARTARIC ACID, ... | | Authors: | Zeng, H, Dong, A, Frommlet, A, Seitova, A, Loppnau, P, Ackloo, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-05 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Co-crystal structure of human DDB1 bound to fragment UB028669
To be published
|
|
8ZKC
 
 | | iron-sulfur cluster transfer protein ApbC | | Descriptor: | GLYCEROL, Iron-sulfur cluster carrier protein, MAGNESIUM ION | | Authors: | Yang, J, Liu, L. | | Deposit date: | 2024-05-16 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of the iron-sulfur cluster transfer protein ApbC from Escherichia coli.
Biochem.Biophys.Res.Commun., 722, 2024
|
|
8Z8X
 
 | | Cryo-EM structure of Thogoto virus polymerase in a transcription initiation conformation | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
2XML
 
 | | Crystal structure of human JMJD2C catalytic domain | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, LYSINE-SPECIFIC DEMETHYLASE 4C, ... | | Authors: | Yue, W.W, Gileadi, C, Krojer, T, Pike, A.C.W, von Delft, F, Ng, S, Carpenter, L, Arrowsmith, C, Weigelt, J, Edwards, A, Bountra, C, Oppermann, U. | | Deposit date: | 2010-07-28 | | Release date: | 2010-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and Evolutionary Basis for the Dual Substrate Selectivity of Human Kdm4 Histone Demethylase Family.
J.Biol.Chem., 286, 2011
|
|
8Y65
 
 | | Cryo-EM structure of human urate transporter GLUT9 bound to substrate urate | | Descriptor: | Solute carrier family 2, facilitated glucose transporter member 9, URIC ACID | | Authors: | Pan, X.J, Shen, Z.L, Xu, L, Huang, G.X.Y. | | Deposit date: | 2024-02-01 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Structural basis for urate recognition and apigenin inhibition of human GLUT9.
Nat Commun, 15, 2024
|
|
8Y66
 
 | | Cryo-EM structure of human urate transporter GLUT9 bound to inhibitor apigenin | | Descriptor: | 5,7-dihydroxy-2-(4-hydroxyphenyl)-4H-chromen-4-one, Solute carrier family 2, facilitated glucose transporter member 9 | | Authors: | Pan, X.J, Shen, Z.L, Xu, L, Huang, G.X.Y. | | Deposit date: | 2024-02-01 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural basis for urate recognition and apigenin inhibition of human GLUT9.
Nat Commun, 15, 2024
|
|
