5ZOH
 
 | |
2CUJ
 
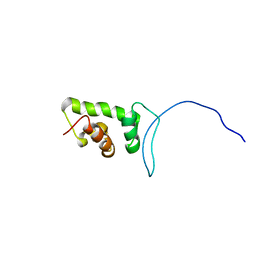 | | Solution structure of SWIRM domain of mouse transcriptional adaptor 2-like | | Descriptor: | transcriptional adaptor 2-like | | Authors: | Yoneyama, M, Umehara, T, Sato, M, Tochio, N, Koshiba, S, Inoue, M, Tanaka, A, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-26 | | Release date: | 2005-11-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Differences of SWIRM Domain Subtypes
J.Mol.Biol., 369, 2007
|
|
1BYA
 
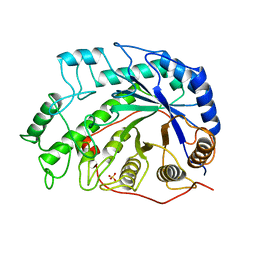 | | CRYSTAL STRUCTURES OF SOYBEAN BETA-AMYLASE REACTED WITH BETA-MALTOSE AND MALTAL: ACTIVE SITE COMPONENTS AND THEIR APPARENT ROLE IN CATALYSIS | | Descriptor: | BETA-AMYLASE, SULFATE ION | | Authors: | Mikami, B, Degano, M, Hehre, E.J, Sacchettini, J.C. | | Deposit date: | 1994-01-25 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of soybean beta-amylase reacted with beta-maltose and maltal: active site components and their apparent roles in catalysis.
Biochemistry, 33, 1994
|
|
1BYB
 
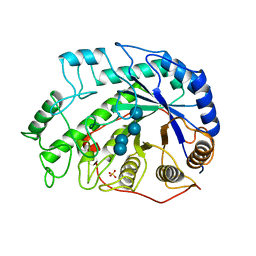 | | CRYSTAL STRUCTURES OF SOYBEAN BETA-AMYLASE REACTED WITH BETA-MALTOSE AND MALTAL: ACTIVE SITE COMPONENTS AND THEIR APPARENT ROLE IN CATALYSIS | | Descriptor: | BETA-AMYLASE, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Mikami, B, Degano, M, Hehre, E.J, Sacchettini, J.C. | | Deposit date: | 1994-01-25 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of soybean beta-amylase reacted with beta-maltose and maltal: active site components and their apparent roles in catalysis.
Biochemistry, 33, 1994
|
|
1BYC
 
 | | CRYSTAL STRUCTURES OF SOYBEAN BETA-AMYLASE REACTED WITH BETA-MALTOSE AND MALTAL: ACTIVE SITE COMPONENTS AND THEIR APPARENT ROLE IN CATALYSIS | | Descriptor: | BETA-AMYLASE, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Mikami, B, Degano, M, Hehre, E.J, Sacchettini, J.C. | | Deposit date: | 1994-01-25 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of soybean beta-amylase reacted with beta-maltose and maltal: active site components and their apparent roles in catalysis.
Biochemistry, 33, 1994
|
|
1BYD
 
 | | CRYSTAL STRUCTURES OF SOYBEAN BETA-AMYLASE REACTED WITH BETA-MALTOSE AND MALTAL: ACTIVE SITE COMPONENTS AND THEIR APPARENT ROLE IN CATALYSIS | | Descriptor: | BETA-AMYLASE, SULFATE ION, alpha-D-glucopyranose-(1-4)-2-deoxy-beta-D-arabino-hexopyranose | | Authors: | Mikami, B, Degano, M, Hehre, E.J, Sacchettini, J.C. | | Deposit date: | 1994-01-25 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of soybean beta-amylase reacted with beta-maltose and maltal: active site components and their apparent roles in catalysis.
Biochemistry, 33, 1994
|
|
1GCU
 
 | |
6AHT
 
 | | Plasmid partitioning protein TubR from Bacillus cereus | | Descriptor: | Conserved hypothetical plasmid protein | | Authors: | Hayashi, I. | | Deposit date: | 2018-08-20 | | Release date: | 2019-06-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cooperative DNA Binding of the Plasmid Partitioning Protein TubR from the Bacillus cereus pXO1 Plasmid.
J.Mol.Biol., 430, 2018
|
|
5YY9
 
 | | Crystal structure of Tandem Tudor Domain of human UHRF1 in complex with LIG1-K126me3 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Ligase 1 | | Authors: | Kori, S, Defossez, P.A, Arita, K. | | Deposit date: | 2017-12-08 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.653 Å) | | Cite: | Structure of the UHRF1 Tandem Tudor Domain Bound to a Methylated Non-histone Protein, LIG1, Reveals Rules for Binding and Regulation.
Structure, 27, 2019
|
|
6AE2
 
 | |
6AE1
 
 | |
5YYA
 
 | | Crystal structure of Tandem Tudor Domain of human UHRF1 | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase UHRF1, SULFATE ION | | Authors: | Kori, S, Defossez, P.A, Arita, K. | | Deposit date: | 2017-12-08 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the UHRF1 Tandem Tudor Domain Bound to a Methylated Non-histone Protein, LIG1, Reveals Rules for Binding and Regulation.
Structure, 27, 2019
|
|
6J13
 
 | | Redox protein from Chlamydomonas reinhardtii | | Descriptor: | 2-cys peroxiredoxin | | Authors: | Charoenwattansatien, R, Zinzius, K, Tanaka, H, Hippler, M, Kurisu, G. | | Deposit date: | 2018-12-27 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Calcium sensing via EF-hand 4 enables thioredoxin activity in the sensor-responder protein calredoxin in the green algaChlamydomonas reinhardtii.
J.Biol.Chem., 295, 2020
|
|
1SRP
 
 | | STRUCTURAL ANALYSIS OF SERRATIA PROTEASE | | Descriptor: | CALCIUM ION, SERRALYSIN, ZINC ION | | Authors: | Hamada, K, Hiramatsu, H, Katsuya, Y, Hata, Y, Katsube, Y. | | Deposit date: | 1994-11-02 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Serratia protease, a zinc-dependent proteinase from Serratia sp. E-15, containing a beta-sheet coil motif at 2.0 A resolution.
J.Biochem.(Tokyo), 119, 1996
|
|
1V6F
 
 | | Solution Structure of Glia Maturation Factor-beta from Mus Musculus | | Descriptor: | glia maturation factor, beta | | Authors: | Goroncy, A.K, Kigawa, T, Koshiba, S, Tomizawa, T, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-29 | | Release date: | 2004-05-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structures of actin depolymerizing factor homology domains.
Protein Sci., 18, 2009
|
|
1WFS
 
 | | Solution Structure of Glia Maturation Factor-gamma from Mus Musculus | | Descriptor: | Glia maturation factor gamma | | Authors: | Goroncy, A.K, Kigawa, T, Koshiba, S, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-26 | | Release date: | 2004-11-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR solution structures of actin depolymerizing factor homology domains.
Protein Sci., 18, 2009
|
|
3VJN
 
 | | Crystal structure of the mutated EGFR kinase domain (G719S/T790M) in complex with AMPPNP. | | Descriptor: | Epidermal growth factor receptor, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Yoshikawa, S, Kukimoto-Niino, M, Shirouzu, M, Semba, K, Yamamoto, T, Yokoyama, S. | | Deposit date: | 2011-10-27 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural basis for the altered drug sensitivities of non-small cell lung cancer-associated mutants of human epidermal growth factor receptor.
Oncogene, 32, 2013
|
|
3UG1
 
 | | Crystal structure of the mutated EGFR kinase domain (G719S/T790M) in the apo form | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Epidermal growth factor receptor | | Authors: | Parker, L.J, Handa, N, Yoshikawa, S, Kukimoto-Niino, M, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2011-11-02 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for the altered drug sensitivities of non-small cell lung cancer-associated mutants of human epidermal growth factor receptor
Oncogene, 32, 2013
|
|
3VJO
 
 | | Crystal structure of the wild-type EGFR kinase domain in complex with AMPPNP. | | Descriptor: | Epidermal growth factor receptor, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Yoshikawa, S, Kukimoto-Niino, M, Shirouzu, M, Semba, K, Yamamoto, T, Yokoyama, S. | | Deposit date: | 2011-10-27 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural basis for the altered drug sensitivities of non-small cell lung cancer-associated mutants of human epidermal growth factor receptor.
Oncogene, 32, 2013
|
|
3UG2
 
 | | Crystal structure of the mutated EGFR kinase domain (G719S/T790M) in complex with gefitinib | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Epidermal growth factor receptor, Gefitinib | | Authors: | Parker, L.J, Handa, N, Yoshikawa, S, Kukimoto-Niino, M, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2011-11-02 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the altered drug sensitivities of non-small cell lung cancer-associated mutants of human epidermal growth factor receptor
Oncogene, 32, 2013
|
|
2ACG
 
 | | ACANTHAMOEBA CASTELLANII PROFILIN II | | Descriptor: | PROFILIN II | | Authors: | Fedorov, A.A, Magnus, K.A, Graupe, M.H, Lattman, E.E, Pollard, T.D, Almo, S.C. | | Deposit date: | 1994-08-30 | | Release date: | 1994-11-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structures of isoforms of the actin-binding protein profilin that differ in their affinity for phosphatidylinositol phosphates.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
6IOY
 
 | | Crystal structure of Porphyromonas gingivalis acetate kinase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Acetate kinase, SULFATE ION | | Authors: | Kezuka, Y, Yoshida, Y, Nonaka, T. | | Deposit date: | 2018-10-31 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Characterization of the phosphotransacetylase-acetate kinase pathway for ATP production inPorphyromonas gingivalis.
J Oral Microbiol, 11, 2019
|
|
6IOW
 
 | |
6INZ
 
 | | Crystal structure of solute-binding protein complexed with unsaturated hyaluronan disaccharide | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Extracellular solute-binding protein family 1, ... | | Authors: | Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2018-10-29 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.289 Å) | | Cite: | Substrate recognition by bacterial solute-binding protein is responsible for import of extracellular hyaluronan and chondroitin sulfate from the animal host.
Biosci.Biotechnol.Biochem., 83, 2019
|
|
6IOX
 
 | |
