4MSE
 
 | | Crystal structure of PDE10A2 with fragment ZT1597 (2-({[(2S)-2-methyl-2,3-dihydro-1,3-benzothiazol-5-yl]oxy}methyl)quinoline) | | Descriptor: | 2-{[(2-methyl-1,3-benzothiazol-5-yl)oxy]methyl}quinoline, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-09-18 | | Release date: | 2014-05-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Identification and optimization of PDE10A inhibitors using fragment-based screening by nanocalorimetry and X-ray crystallography.
J Biomol Screen, 19, 2014
|
|
4MRZ
 
 | | Crystal structure of PDE10A2 with fragment ZT0429 (4-methyl-3-nitropyridin-2-amine) | | Descriptor: | 4-methyl-3-nitropyridin-2-amine, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-09-17 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Identification and optimization of PDE10A inhibitors using fragment-based screening by nanocalorimetry and X-ray crystallography.
J Biomol Screen, 19, 2014
|
|
4MSA
 
 | | Crystal structure of PDE10A2 with fragment ZT0449 (5-nitro-1H-benzimidazole) | | Descriptor: | 5-nitro-1H-benzimidazole, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-09-18 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Identification and optimization of PDE10A inhibitors using fragment-based screening by nanocalorimetry and X-ray crystallography.
J Biomol Screen, 19, 2014
|
|
4MS0
 
 | | Crystal structure of PDE10A2 with fragment ZT0443 (6-chloropyrimidine-2,4-diamine) | | Descriptor: | 6-chloropyrimidine-2,4-diamine, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-09-18 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Identification and optimization of PDE10A inhibitors using fragment-based screening by nanocalorimetry and X-ray crystallography.
J Biomol Screen, 19, 2014
|
|
4NI0
 
 | | Quaternary R3 CO-liganded hemoglobin structure in complex with a thiol containing compound | | Descriptor: | 5-[(2S)-2,3-dihydro-1,4-benzodioxin-2-yl]-2,4-dihydro-3H-1,2,4-triazole-3-thione, CARBON MONOXIDE, Hemoglobin subunit alpha, ... | | Authors: | Safo, M.K, Meadows, J, Ko, T.-P, Nakagawa, A, Zapol, W. | | Deposit date: | 2013-11-05 | | Release date: | 2014-08-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Identification of a Small Molecule that Increases Hemoglobin Oxygen Affinity and Reduces SS Erythrocyte Sickling.
Acs Chem.Biol., 9, 2014
|
|
4NI1
 
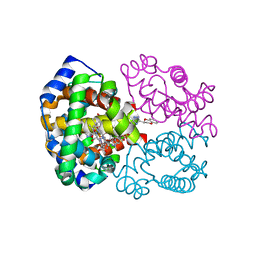 | | Quaternary R CO-liganded hemoglobin structure in complex with a thiol containing compound | | Descriptor: | 5-[(2R)-2,3-dihydro-1,4-benzodioxin-2-yl]-2,4-dihydro-3H-1,2,4-triazole-3-thione, CARBON MONOXIDE, Hemoglobin subunit alpha, ... | | Authors: | Safo, M.K, Meadows, J, Ko, T.-P, Nakagawa, A, Zapol, W. | | Deposit date: | 2013-11-05 | | Release date: | 2014-09-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of a Small Molecule that Increases Hemoglobin Oxygen Affinity and Reduces SS Erythrocyte Sickling.
Acs Chem.Biol., 9, 2014
|
|
4MSN
 
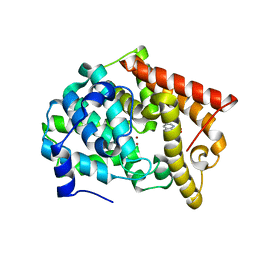 | | Crystal structure of PDE10A2 with fragment ZT0451 (8-nitroquinoline) | | Descriptor: | 8-nitroquinoline, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-09-18 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification and optimization of PDE10A inhibitors using fragment-based screening by nanocalorimetry and X-ray crystallography.
J Biomol Screen, 19, 2014
|
|
1VKX
 
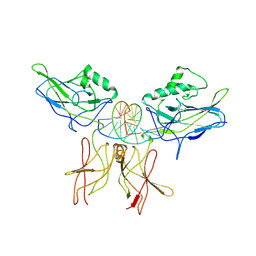 | | CRYSTAL STRUCTURE OF THE NFKB P50/P65 HETERODIMER COMPLEXED TO THE IMMUNOGLOBULIN KB DNA | | Descriptor: | DNA (5'-D(*AP*GP*GP*AP*AP*AP*GP*TP*CP*CP*CP*C)-3'), DNA (5'-D(*TP*GP*GP*GP*GP*AP*CP*TP*TP*TP*CP*C)-3'), PROTEIN (NF-KAPPA B P50 SUBUNIT), ... | | Authors: | Chen, F, Huang, D.B, Ghosh, G. | | Deposit date: | 1997-09-17 | | Release date: | 1998-12-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of p50/p65 heterodimer of transcription factor NF-kappaB bound to DNA.
Nature, 391, 1998
|
|
1ANV
 
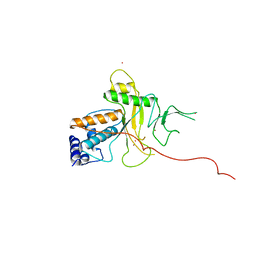 | | ADENOVIRUS 5 DBP/URANYL FLUORIDE SOAK | | Descriptor: | ADENOVIRUS SINGLE-STRANDED DNA-BINDING PROTEIN, URANYL (VI) ION, ZINC ION | | Authors: | Kanellopoulos, P.N, Tsernoglou, D, Van Der Vliet, P.C, Tucker, P.A. | | Deposit date: | 1996-03-14 | | Release date: | 1997-02-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Conformational change of the adenovirus DNA-binding protein induced by soaking crystals with K3UO2F5 solutions.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1ADU
 
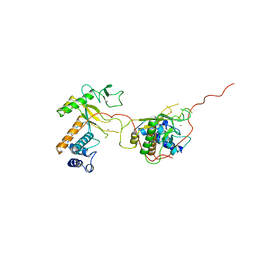 | | EARLY E2A DNA-BINDING PROTEIN | | Descriptor: | ADENOVIRUS SINGLE-STRANDED DNA-BINDING PROTEIN, ZINC ION | | Authors: | Tucker, P.A, Kanellopoulos, P.N, Tsernoglou, D, Van Der Vliet, P.C. | | Deposit date: | 1995-05-11 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Alternative arrangements of the protein chain are possible for the adenovirus single-stranded DNA binding protein.
J.Mol.Biol., 257, 1996
|
|
1ADV
 
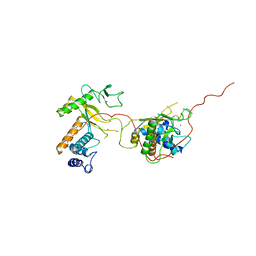 | | EARLY E2A DNA-BINDING PROTEIN | | Descriptor: | ADENOVIRUS SINGLE-STRANDED DNA-BINDING PROTEIN, ZINC ION | | Authors: | Kanellopoulos, P.N, Tsernoglou, D, Van Der Vliet, P.C, Tucker, P.A. | | Deposit date: | 1995-05-12 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Alternative arrangements of the protein chain are possible for the adenovirus single-stranded DNA binding protein.
J.Mol.Biol., 257, 1996
|
|
1NST
 
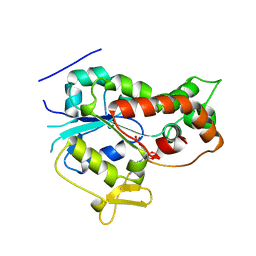 | |
1EIT
 
 | | NMR STUDY OF MU-AGATOXIN | | Descriptor: | MU-AGATOXIN-I | | Authors: | Omecinsky, D.O, Reily, M.D. | | Deposit date: | 1995-12-14 | | Release date: | 1996-04-03 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure analysis of mu-agatoxins: further evidence for common motifs among neurotoxins with diverse ion channel specificities.
Biochemistry, 35, 1996
|
|
4LVW
 
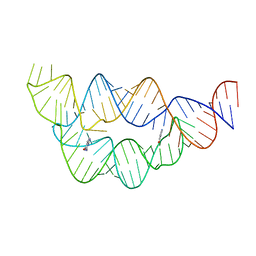 | |
3U0S
 
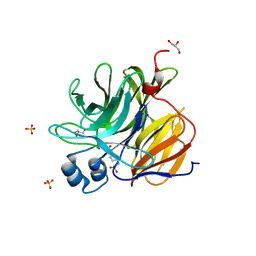 | | Crystal Structure of an Enzyme Redesigned Through Multiplayer Online Gaming: CE6 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Diisopropyl-fluorophosphatase, GLYCEROL, ... | | Authors: | Bale, J.B, Shen, B.W, Stoddard, B.L. | | Deposit date: | 2011-09-29 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Increased Diels-Alderase activity through backbone remodeling guided by Foldit players.
Nat.Biotechnol., 30, 2012
|
|
1RYQ
 
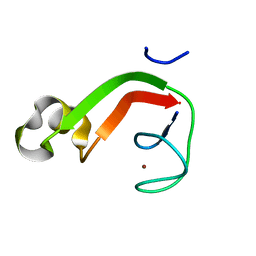 | | Putative DNA-directed RNA polymerase, subunit e'' from Pyrococcus Furiosus Pfu-263306-001 | | Descriptor: | DNA-directed RNA polymerase, subunit e'', ZINC ION | | Authors: | Liu, Z.-J, Chen, L, Tempel, W, Shah, A, Arendall III, W.B, Rose, J.P, Brereton, P.S, Izumi, M, Jenney Jr, F.E, Lee, H.S, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-12-22 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Parameter-space screening: a powerful tool for high-throughput crystal structure determination.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
4WRN
 
 | | Crystal structure of the polymerization region of human uromodulin/Tamm-Horsfall protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Maltose-binding periplasmic protein,Uromodulin, ZINC ION, ... | | Authors: | Bokhove, M, De Sanctis, D, Jovine, L. | | Deposit date: | 2014-10-24 | | Release date: | 2016-01-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A structured interdomain linker directs self-polymerization of human uromodulin.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1JES
 
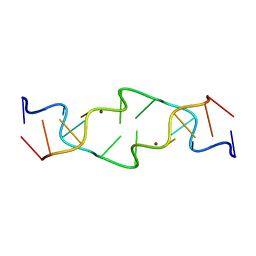 | | Crystal Structure of a Copper-Mediated Base Pair in DNA | | Descriptor: | 5'-D(*CP*GP*CP*GP*(DPY)P*AP*TP*(DRP)P*CP*GP*CP*G)-3', COPPER (II) ION | | Authors: | Atwell, S, Meggers, E, Spraggon, G, Schultz, P.G. | | Deposit date: | 2001-06-18 | | Release date: | 2001-11-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a Copper-Mediated Base Pair in DNA
J.Am.Chem.Soc., 123, 2001
|
|
4LVX
 
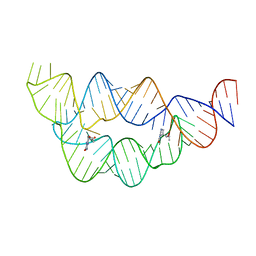 | |
4LVZ
 
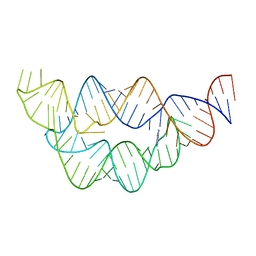 | |
4LVY
 
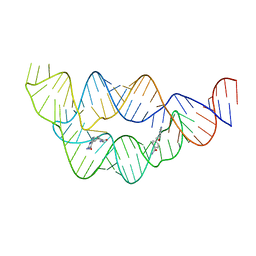 | | Structure of the THF riboswitch bound to pemetrexed | | Descriptor: | 2-{4-[2-(2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDIN-5-YL)-ETHYL]-BENZOYLAMINO}-PENTANEDIOIC ACID, THF riboswitch | | Authors: | Trausch, J.J, Batey, R.T. | | Deposit date: | 2013-07-26 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Disconnect between High-Affinity Binding and Efficient Regulation by Antifolates and Purines in the Tetrahydrofolate Riboswitch.
Chem.Biol., 21, 2014
|
|
4LVV
 
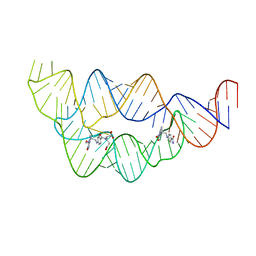 | | Structure of the THF riboswitch | | Descriptor: | N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, THF riboswitch | | Authors: | Trausch, J.J, Batey, R.T. | | Deposit date: | 2013-07-26 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Disconnect between High-Affinity Binding and Efficient Regulation by Antifolates and Purines in the Tetrahydrofolate Riboswitch.
Chem.Biol., 21, 2014
|
|
4LW0
 
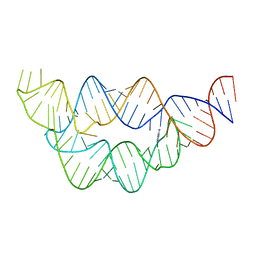 | | Structure of the THF riboswitch bound to adenine | | Descriptor: | ADENINE, THF riboswitch | | Authors: | Trausch, J.J, Batey, R.T. | | Deposit date: | 2013-07-26 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.889 Å) | | Cite: | A Disconnect between High-Affinity Binding and Efficient Regulation by Antifolates and Purines in the Tetrahydrofolate Riboswitch.
Chem.Biol., 21, 2014
|
|
1VK1
 
 | | Conserved hypothetical protein from Pyrococcus furiosus Pfu-392566-001 | | Descriptor: | Conserved hypothetical protein, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Shah, A, Liu, Z.J, Tempel, W, Chen, L, Lee, D, Yang, H, Chang, J, Zhao, M, Ng, J, Rose, J, Brereton, P.S, Izumi, M, Jenney Jr, F.E, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-04-13 | | Release date: | 2004-08-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | (NZ)CH...O contacts assist crystallization of a ParB-like nuclease.
Bmc Struct.Biol., 7, 2007
|
|
8U81
 
 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: State A From Composite RELION Multi-body Refinement Map | | Descriptor: | BTB/POZ domain-containing protein KCTD5, Cullin-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | Deposit date: | 2023-09-15 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structure and dynamics of a pentameric KCTD5/CUL3/G beta gamma E3 ubiquitin ligase complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
