3QVR
 
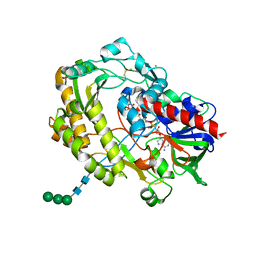 | | Crystal structure of glucose oxidase for space group P3121 at 1.3 A resolution. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kommoju, P, Chen, Z, Bruckner, R.C, Mathews, F.S, Jorns, M.S. | | Deposit date: | 2011-02-25 | | Release date: | 2011-06-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Probing oxygen activation sites in two flavoprotein oxidases using chloride as an oxygen surrogate.
Biochemistry, 50, 2011
|
|
3QVP
 
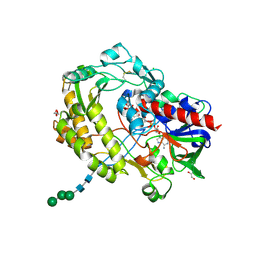 | | Crystal structure of glucose oxidase for space group C2221 at 1.2 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kommoju, P, Chen, Z, Bruckner, R.C, Mathews, F.S, Jorns, M.S. | | Deposit date: | 2011-02-25 | | Release date: | 2011-06-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Probing oxygen activation sites in two flavoprotein oxidases using chloride as an oxygen surrogate.
Biochemistry, 50, 2011
|
|
1QGE
 
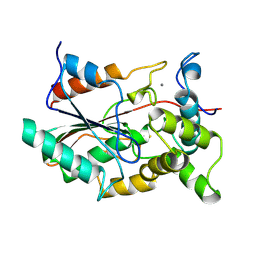 | | NEW CRYSTAL FORM OF PSEUDOMONAS GLUMAE (FORMERLY CHROMOBACTERIUM VISCOSUM ATCC 6918) LIPASE | | Descriptor: | CALCIUM ION, PROTEIN (TRIACYLGLYCEROL HYDROLASE) | | Authors: | Lang, D.A, Stadler, P, Kovacs, A, Paltauf, F, Dijkstra, B.W. | | Deposit date: | 1999-04-27 | | Release date: | 1999-05-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Kinetic Investigations of Enantiomeric Binding Mode of Subclass I Lipases from the Family of Pseudomonadaceae
To be Published
|
|
5WYM
 
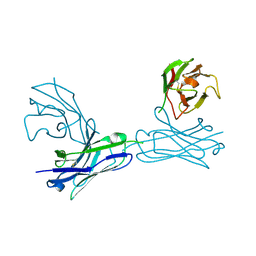 | | Crystal structure of an anti-connexin26 scFv | | Descriptor: | anti-connexin26 scFv,Ig heavy chain,Linker,anti-connexin26 scFv,Ig light chain | | Authors: | Li, S, Xu, L. | | Deposit date: | 2017-01-13 | | Release date: | 2018-01-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Design and Characterization of a Human Monoclonal Antibody that Modulates Mutant Connexin 26 Hemichannels Implicated in Deafness and Skin Disorders
Front Mol Neurosci, 10, 2017
|
|
1B3K
 
 | | Plasminogen activator inhibitor-1 | | Descriptor: | PLASMINOGEN ACTIVATOR INHIBITOR-1 | | Authors: | Sharp, A.M, Stein, P.E, Pannu, N.S, Read, R.J. | | Deposit date: | 1998-12-11 | | Release date: | 1999-12-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The active conformation of plasminogen activator inhibitor 1, a target for drugs to control fibrinolysis and cell adhesion.
Structure Fold.Des., 7, 1999
|
|
1HNY
 
 | |
3LMX
 
 | | Tyrosine 447 of Protocatechuate 34,-Dioxygenase Controls Efficient Progress Through Catalysis | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Purpero, V.M, Lipscomb, J.D, Shi, K. | | Deposit date: | 2010-02-01 | | Release date: | 2011-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Tyrosine 447 of Protocatechuate 34,-Dioxygenase Controls Efficient Progress Through Catalysis
To be Published
|
|
3LN0
 
 | | Structure of compound 5c-S bound at the active site of COX-2 | | Descriptor: | (2S)-6,8-dichloro-2-(trifluoromethyl)-2H-chromene-3-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kiefer, J.R, Kurumbail, R.G, Stallings, W.C, Pawlitz, J.L. | | Deposit date: | 2010-02-01 | | Release date: | 2010-10-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The novel benzopyran class of selective cyclooxygenase-2 inhibitors. Part 2: The second clinical candidate having a shorter and favorable human half-life.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3LKT
 
 | |
1GPE
 
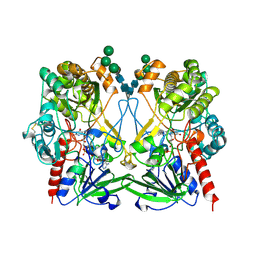 | | GLUCOSE OXIDASE FROM PENICILLIUM AMAGASAKIENSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Hendle, J, Kalisz, H.M, Hecht, H.J. | | Deposit date: | 1999-03-24 | | Release date: | 1999-05-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 and 1.9 A resolution structures of the Penicillium amagasakiense and Aspergillus niger glucose oxidases as a basis for modelling substrate complexes.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
3LN1
 
 | | Structure of celecoxib bound at the COX-2 active site | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[5-(4-METHYLPHENYL)-3-(TRIFLUOROMETHYL)-1H-PYRAZOL-1-YL]BENZENESULFONAMIDE, ... | | Authors: | Kiefer, J.R, Kurumbail, R.G, Stallings, W.C, Pawlitz, J.L. | | Deposit date: | 2010-02-01 | | Release date: | 2010-10-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The novel benzopyran class of selective cyclooxygenase-2 inhibitors. Part 2: The second clinical candidate having a shorter and favorable human half-life.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3M35
 
 | |
3M36
 
 | |
3M37
 
 | |
5Q0V
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | (2S)-2-[2-(4-chlorophenyl)-5,6-difluoro-1H-benzimidazol-1-yl]-2-cyclohexyl-N-(2-fluorophenyl)acetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q1A
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | (2S)-2-cyclohexyl-2-[2-(2,4-dimethoxyphenyl)-1H-benzimidazol-1-yl]-N-(2,6-dimethylphenyl)acetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q19
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | (2S)-N,2-dicyclohexyl-2-[2-(2,4-dimethoxyphenyl)-1H-benzimidazol-1-yl]acetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0R
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3, N,1-dibenzyl-6-[(2-fluorophenyl)sulfonyl]-4,5,6,7-tetrahydro-1H-pyrrolo[2,3-c]pyridine-2-carboxamide | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0Z
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3, ethyl (5S)-3-(3,4-difluorobenzene-1-carbonyl)-1,1-dimethyl-1,2,3,4,5,6-hexahydroazepino[4,5-b]indole-5-carboxylate | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q1I
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 3-(2-chlorophenyl)-N-[(1R)-1-(naphthalen-2-yl)ethyl]-5-(propan-2-yl)-1,2-oxazole-4-carboxamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
1SPS
 
 | |
2GAJ
 
 | |
4J98
 
 | |
4J95
 
 | |
4J97
 
 | |
