7CDJ
 
 | | Crystal structure of SARS-CoV-2 antibody P2C-1A3 with RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P2C-1A3 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Ge, J, Wang, R. | | Deposit date: | 2020-06-19 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.396 Å) | | Cite: | Antibody neutralization of SARS-CoV-2 through ACE2 receptor mimicry.
Nat Commun, 12, 2021
|
|
7F4G
 
 | | Structure of RPAP2-bound RNA polymerase II | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Chen, X, Qi, Y, Wang, X, Li, J, Zhao, D, Xu, Y. | | Deposit date: | 2021-06-18 | | Release date: | 2021-07-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | RPAP2 regulates a transcription initiation checkpoint by inhibiting assembly of pre-initiation complex.
Cell Rep, 39, 2022
|
|
7FEE
 
 | | Crystal structure of the allosteric modulator ZCZ011 binding to CP55940-bound cannabinoid receptor 1 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-[(1R,2R,5R)-5-hydroxy-2-(3-hydroxypropyl)cyclohexyl]-5-(2-methyloctan-2-yl)phenol, 6-methyl-3-[(1S)-2-nitro-1-thiophen-2-yl-ethyl]-2-phenyl-1H-indole, ... | | Authors: | Wang, X, Zhao, C, Shao, Z. | | Deposit date: | 2021-07-19 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular mechanism of allosteric modulation for the cannabinoid receptor CB1.
Nat.Chem.Biol., 18, 2022
|
|
7CWU
 
 | | SARS-CoV-2 spike proteins trimer in complex with P17 and FC05 Fabs cocktail | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, X, Wang, N. | | Deposit date: | 2020-08-31 | | Release date: | 2020-12-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure-based development of human antibody cocktails against SARS-CoV-2.
Cell Res., 31, 2021
|
|
7DHD
 
 | | Vibrio vulnificus Wzb | | Descriptor: | CHLORIDE ION, Protein-tyrosine-phosphatase | | Authors: | Ma, Q, Wang, X. | | Deposit date: | 2020-11-14 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Wzb of Vibrio vulnificus represents a new group of low-molecular-weight protein tyrosine phosphatases with a unique insertion in the W-loop.
J.Biol.Chem., 296, 2021
|
|
7DHF
 
 | | Vibrio vulnificus Wzb in complex with benzylphosphonate | | Descriptor: | GLYCEROL, PHOSPHATE ION, Protein-tyrosine-phosphatase | | Authors: | Ma, Q, Wang, X. | | Deposit date: | 2020-11-14 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.211 Å) | | Cite: | Wzb of Vibrio vulnificus represents a new group of low-molecular-weight protein tyrosine phosphatases with a unique insertion in the W-loop.
J.Biol.Chem., 296, 2021
|
|
7DHE
 
 | | Vibrio vulnificus Wzb in complex with benzylphosphonate | | Descriptor: | Protein-tyrosine-phosphatase, benzylphosphonic acid | | Authors: | Ma, Q, Wang, X. | | Deposit date: | 2020-11-14 | | Release date: | 2021-01-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Wzb of Vibrio vulnificus represents a new group of low-molecular-weight protein tyrosine phosphatases with a unique insertion in the W-loop.
J.Biol.Chem., 296, 2021
|
|
7CN8
 
 | | Cryo-EM structure of PCoV_GX spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein, ... | | Authors: | Wang, X, Yu, J, Zhang, S, Qiao, S, Zeng, J, Tian, L. | | Deposit date: | 2020-07-30 | | Release date: | 2021-03-03 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Bat and pangolin coronavirus spike glycoprotein structures provide insights into SARS-CoV-2 evolution.
Nat Commun, 12, 2021
|
|
7CN4
 
 | | Cryo-EM structure of bat RaTG13 spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Zhang, S, Qiao, S, Yu, J, Zeng, J, Tian, L. | | Deposit date: | 2020-07-30 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Bat and pangolin coronavirus spike glycoprotein structures provide insights into SARS-CoV-2 evolution.
Nat Commun, 12, 2021
|
|
7FCC
 
 | | IL-1RAcPb TIR domain | | Descriptor: | Isoform 4 of Interleukin-1 receptor accessory protein | | Authors: | Wang, X, Zhou, J. | | Deposit date: | 2021-07-14 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.144 Å) | | Cite: | Structural basis of the IL-1 receptor TIR domain-mediated IL-1 signaling
Iscience, 25, 2022
|
|
7FCJ
 
 | | Zebrafish SIGIRR TIR domain mutant - C299S | | Descriptor: | SIGIRR protein | | Authors: | Wang, X, Zhou, J. | | Deposit date: | 2021-07-15 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural basis of the IL-1 receptor TIR domain-mediated IL-1 signaling
Iscience, 25, 2022
|
|
7FCH
 
 | | IL-18Rbeta TIR domain | | Descriptor: | Interleukin-18 receptor accessory protein | | Authors: | Wang, X, Zhou, J. | | Deposit date: | 2021-07-14 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.883 Å) | | Cite: | Structural basis of the IL-1 receptor TIR domain-mediated IL-1 signaling
Iscience, 25, 2022
|
|
7FD3
 
 | | IL-1RAPL2 TIR domain | | Descriptor: | X-linked interleukin-1 receptor accessory protein-like 2 | | Authors: | Wang, X, Zhou, J. | | Deposit date: | 2021-07-15 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural basis of the IL-1 receptor TIR domain-mediated IL-1 signaling
Iscience, 25, 2022
|
|
7FCL
 
 | | Zebrafish SIGIRR TIR domain | | Descriptor: | SIGIRR protein | | Authors: | Wang, X, Zhou, J. | | Deposit date: | 2021-07-15 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structural basis of the IL-1 receptor TIR domain-mediated IL-1 signaling
Iscience, 25, 2022
|
|
7CVN
 
 | | The N-arylsulfonyl-indole-2-carboxamide-based inhibitors against fructose-1,6-bisphosphatase | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 4-(3-acetamidophenyl)-N-(4-methoxyphenyl)sulfonyl-7-nitro-1H-indole-2-carboxamide, Fructose-1,6-bisphosphatase 1 | | Authors: | Wang, X, Zhou, J, Xu, B. | | Deposit date: | 2020-08-26 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Design,synthesis,biological evaluation and binding mode analysis of 7-nitro-indole-N-acylarylsulfonamide-based fructose-1,6-bisphosphatase inhibitors
Chinese journal of medicinal chemistry, 30, 2020
|
|
6U98
 
 | | Hsp90a NTD K58R bound reversibly to sulfonyl fluoride 6 | | Descriptor: | 3-{[(3R)-3-({6-amino-8-[(6-iodo-2H-1,3-benzodioxol-5-yl)sulfanyl]-9H-purin-9-yl}methyl)piperidin-1-yl]methyl}benzene-1-sulfonyl fluoride, Heat shock protein HSP 90-alpha, POTASSIUM ION, ... | | Authors: | Cuesta, A, Wan, X, Taunton, J. | | Deposit date: | 2019-09-06 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Ligand Conformational Bias Drives Enantioselective Modification of a Surface-Exposed Lysine on Hsp90.
J.Am.Chem.Soc., 142, 2020
|
|
6U9B
 
 | | Hsp90a NTD covalently bound to sulfonyl fluoride 5 at K58 | | Descriptor: | 3-{[(3S)-3-({6-amino-8-[(6-iodo-2H-1,3-benzodioxol-5-yl)sulfanyl]-9H-purin-9-yl}methyl)piperidin-1-yl]methyl}benzene-1-sulfonyl fluoride, Heat shock protein HSP 90-alpha | | Authors: | Cuesta, A, Wan, X, Taunton, J. | | Deposit date: | 2019-09-07 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Ligand Conformational Bias Drives Enantioselective Modification of a Surface-Exposed Lysine on Hsp90.
J.Am.Chem.Soc., 142, 2020
|
|
6U99
 
 | | Hsp90a NTD covalently bound to sulfonyl fluoride probe 1 at K58 | | Descriptor: | 3-{[(3-{6-amino-8-[(6-iodo-2H-1,3-benzodioxol-5-yl)sulfanyl]-9H-purin-9-yl}propyl)amino]methyl}benzene-1-sulfinic acid, Heat shock protein HSP 90-alpha | | Authors: | Cuesta, A, Wan, X, Taunton, J. | | Deposit date: | 2019-09-06 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Ligand Conformational Bias Drives Enantioselective Modification of a Surface-Exposed Lysine on Hsp90.
J.Am.Chem.Soc., 142, 2020
|
|
5GWE
 
 | | cytochrome P450 CREJ | | Descriptor: | (4-methylphenyl) dihydrogen phosphate, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Dong, S, liu, X, Wang, X, Feng, Y. | | Deposit date: | 2016-09-11 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selective oxidation of aliphatic C-H bonds in alkylphenols by a chemomimetic biocatalytic system
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2W31
 
 | | globin domain of Geobacter sulfurreducens globin-coupled sensor | | Descriptor: | GLOBIN, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Pesce, A, Thijs, L, Nardini, M, Desmet, F, Sisinni, L, Gourlay, L, Bolli, A, Coletta, M, Van Doorslaer, S, Wan, X, Alam, M, Ascenzi, P, Moens, L, Bolognesi, M, Dewilde, S. | | Deposit date: | 2008-11-05 | | Release date: | 2009-01-13 | | Last modified: | 2019-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Hise11 and Hisf8 Provide Bis-Histidyl Heme Hexa-Coordination in the Globin Domain of Geobacter Sulfurreducens Globin-Coupled Sensor.
J.Mol.Biol., 386, 2009
|
|
4RC2
 
 | |
6KZI
 
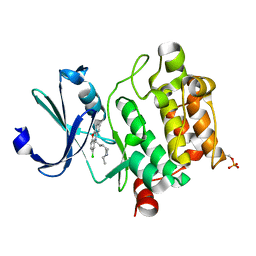 | | Crystal structure of Ser/Thr kinase Pim1 in complex with thioridazine derivatives | | Descriptor: | 4-(2-chloro-10H-phenoxazin-10-yl)-N,N-diethylbutan-1-amine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Wan, X, Li, W, Xie, Y, Huang, N. | | Deposit date: | 2019-09-24 | | Release date: | 2020-03-04 | | Last modified: | 2021-03-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
6U9A
 
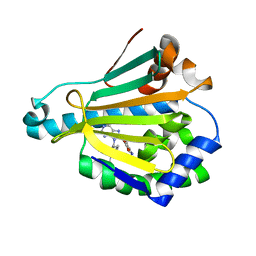 | | Hsp90a NTD K58R bound reversibly to sulfonyl fluoride 5 | | Descriptor: | 3-{[(3S)-3-({6-amino-8-[(6-iodo-2H-1,3-benzodioxol-5-yl)sulfanyl]-9H-purin-9-yl}methyl)piperidin-1-yl]methyl}benzene-1-sulfonyl fluoride, Heat shock protein HSP 90-alpha | | Authors: | Cuesta, A, Wan, X, Taunton, J. | | Deposit date: | 2019-09-07 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Ligand Conformational Bias Drives Enantioselective Modification of a Surface-Exposed Lysine on Hsp90.
J.Am.Chem.Soc., 142, 2020
|
|
4IAA
 
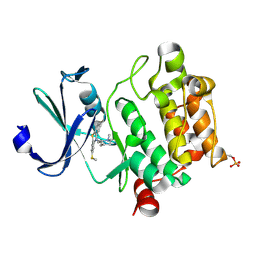 | | Crystal structure of Ser/Thr kinase Pim1 in complex with thioridazine | | Descriptor: | 10-{2-[(2R)-1-methylpiperidin-2-yl]ethyl}-2-(methylsulfanyl)-10H-phenothiazine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Wan, X, Li, W, Xie, Y, Huang, N. | | Deposit date: | 2012-12-06 | | Release date: | 2013-12-18 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of Ser/Thr kinase Pim1 in complex with thioridazine
To be Published
|
|
5C1Q
 
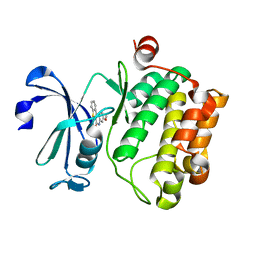 | | Serine/threonine-protein kinase pim-1 | | Descriptor: | 3-methoxy[1]benzothieno[2,3-c]quinolin-6(5H)-one, Serine/threonine-protein kinase pim-1 | | Authors: | Li, W, Wan, X, Huang, N. | | Deposit date: | 2015-06-15 | | Release date: | 2015-12-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Serine/threonine-protein kinase pim-1
To Be Published
|
|
