7WYM
 
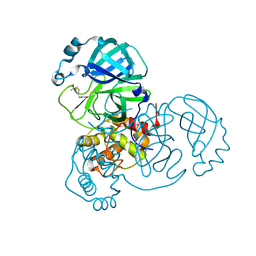 | | Structure of the SARS-COV-2 main protease with 337 inhibitor | | Descriptor: | 3C-like proteinase nsp5, N-methyl-N-[[4-(trifluoromethyl)-1,3-thiazol-2-yl]methyl]prop-2-enamide | | Authors: | Qin, B, Hou, P, Gao, X, Cui, S. | | Deposit date: | 2022-02-16 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Acrylamide fragment inhibitors that induce unprecedented conformational distortions in enterovirus 71 3C and SARS-CoV-2 main protease.
Acta Pharm Sin B, 12, 2022
|
|
7WYP
 
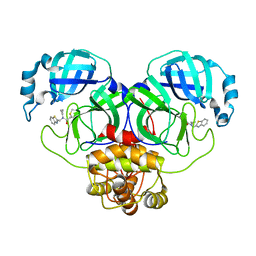 | | Structure of the SARS-COV-2 main protease with EN102 inhibitor | | Descriptor: | 3C-like proteinase, N-(1,3-benzothiazol-2-ylmethyl)-N-cyclopropyl-prop-2-enamide | | Authors: | Qin, B, Hou, P, Gao, X, Cui, S. | | Deposit date: | 2022-02-16 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Acrylamide fragment inhibitors that induce unprecedented conformational distortions in enterovirus 71 3C and SARS-CoV-2 main protease.
Acta Pharm Sin B, 12, 2022
|
|
5X5O
 
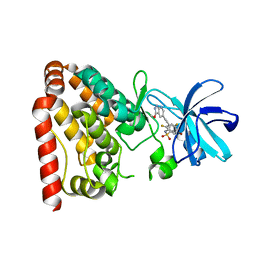 | | Crystal structure of ZAK in complex with compound D2829 | | Descriptor: | Mitogen-activated protein kinase kinase kinase MLT, N-[2,4-bis(fluoranyl)-3-[2-(3-methoxy-1H-pyrazolo[3,4-b]pyridin-5-yl)ethynyl]phenyl]-3-bromanyl-benzenesulfonamide | | Authors: | Dai, Y.B, Zhao, P, Yun, C.H. | | Deposit date: | 2017-02-17 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.868 Å) | | Cite: | Structure Based Design of N-(3-((1H-Pyrazolo[3,4-b]pyridin-5-yl)ethynyl)benzenesulfonamides as Selective Leucine-Zipper and Sterile-alpha Motif Kinase (ZAK) Inhibitors.
J. Med. Chem., 60, 2017
|
|
6JRK
 
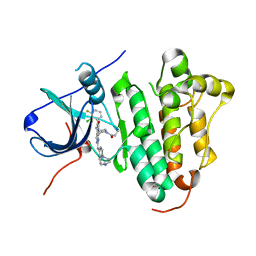 | | The structure of co-crystals of 8r-B-EGFR WT complex | | Descriptor: | 6-(2-chloranyl-3-fluoranyl-phenyl)-5-methyl-2-[[3-methyl-4-(4-methylpiperazin-1-yl)phenyl]amino]-8-[(3S)-1-propanoylpiperidin-3-yl]pyrido[2,3-d]pyrimidin-7-one, Epidermal growth factor receptor | | Authors: | Zhu, S.J, Yun, C.H. | | Deposit date: | 2019-04-04 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Structure-Based Design of 5-Methylpyrimidopyridone Derivatives as New Wild-Type Sparing Inhibitors of the Epidermal Growth Factor Receptor Triple Mutant (EGFRL858R/T790M/C797S).
J.Med.Chem., 62, 2019
|
|
6JRJ
 
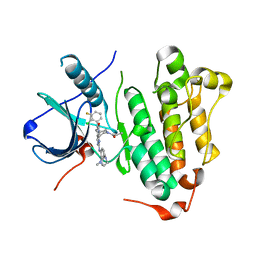 | | The structure of co-crystals of 8r-B-EGFR T790M/C797S complex | | Descriptor: | 6-(2-chloranyl-3-fluoranyl-phenyl)-5-methyl-2-[[3-methyl-4-(4-methylpiperazin-1-yl)phenyl]amino]-8-[(3S)-1-propanoylpiperidin-3-yl]pyrido[2,3-d]pyrimidin-7-one, Epidermal growth factor receptor | | Authors: | Zhu, S.J, Yun, C.H. | | Deposit date: | 2019-04-04 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.943 Å) | | Cite: | Structure-Based Design of 5-Methylpyrimidopyridone Derivatives as New Wild-Type Sparing Inhibitors of the Epidermal Growth Factor Receptor Triple Mutant (EGFRL858R/T790M/C797S).
J.Med.Chem., 62, 2019
|
|
6JRX
 
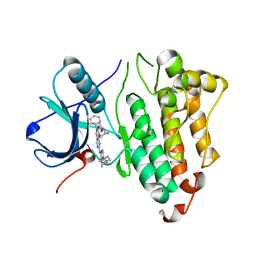 | | EGFR T790M/C797S in complex with compound 6i | | Descriptor: | Epidermal growth factor receptor, N-{trans-4-[3-(2-chlorophenyl)-7-{[3-methyl-4-(4-methylpiperazin-1-yl)phenyl]amino}-2-oxo-3,4-dihydropyrimido[4,5-d]pyrimidin-1(2H)-yl]cyclohexyl}propanamide | | Authors: | Zhu, S.J, Yun, C.H. | | Deposit date: | 2019-04-06 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Design, Synthesis, and Structure-Activity Relationships of 1,2,3-Triazole Benzenesulfonamides as New Selective Leucine-Zipper and Sterile-alpha Motif Kinase (ZAK) Inhibitors.
J.Med.Chem., 63, 2020
|
|
6JUU
 
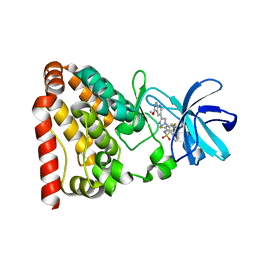 | | Crystal structure of ZAK in complex with compound 6r | | Descriptor: | Mitogen-activated protein kinase kinase kinase MLT, ~{N}-[2,4-bis(fluoranyl)-3-[4-(3-methoxy-1~{H}-pyrazolo[3,4-b]pyridin-5-yl)-1,2,3-triazol-1-yl]phenyl]naphthalene-1-sulfonamide | | Authors: | Kong, L.L, Yun, C.H. | | Deposit date: | 2019-04-15 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Design, Synthesis, and Structure-Activity Relationships of 1,2,3-Triazole Benzenesulfonamides as New Selective Leucine-Zipper and Sterile-alpha Motif Kinase (ZAK) Inhibitors.
J.Med.Chem., 63, 2020
|
|
6JUT
 
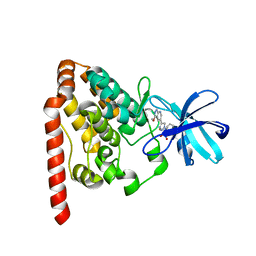 | | Crystal structure of ZAK in complex with compound 6k | | Descriptor: | Mitogen-activated protein kinase kinase kinase MLT, ~{N}-[2,4-bis(fluoranyl)-3-[4-(3-methoxy-1~{H}-pyrazolo[3,4-b]pyridin-5-yl)-1,2,3-triazol-1-yl]phenyl]-3-bromanyl-benzenesulfonamide | | Authors: | Kong, L.L, Yun, C.H. | | Deposit date: | 2019-04-15 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design, Synthesis, and Structure-Activity Relationships of 1,2,3-Triazole Benzenesulfonamides as New Selective Leucine-Zipper and Sterile-alpha Motif Kinase (ZAK) Inhibitors.
J.Med.Chem., 63, 2020
|
|
6HP9
 
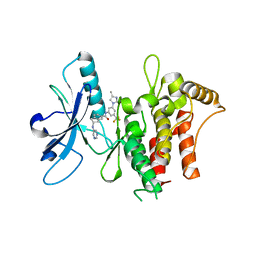 | | Structure of the kinase domain of human DDR1 in complex with a 2-Amino-2,3-Dihydro-1H-Indene-5-Carboxamide-based inhibitor | | Descriptor: | (2~{R})-~{N}-[3-(4-methylimidazol-1-yl)-5-(trifluoromethyl)phenyl]-2-(pyrimidin-5-ylamino)-2,3-dihydro-1~{H}-indene-5-carboxamide, Epithelial discoidin domain-containing receptor 1 | | Authors: | Pinkas, D.M, Fox, A.E, Kupinska, K, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-19 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | 2-Amino-2,3-dihydro-1H-indene-5-carboxamide-Based Discoidin Domain Receptor 1 (DDR1) Inhibitors: Design, Synthesis, and in Vivo Antipancreatic Cancer Efficacy.
J.Med.Chem., 62, 2019
|
|
3CW9
 
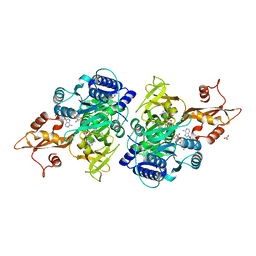 | | 4-Chlorobenzoyl-CoA Ligase/Synthetase in the Thioester-forming Conformation, bound to 4-chlorophenacyl-CoA | | Descriptor: | 1,2-ETHANEDIOL, 4-Chlorophenacyl-coenzyme A, 4-chlorobenzoyl CoA ligase, ... | | Authors: | Reger, A.S, Cao, J, Wu, R, Dunaway-Mariano, D, Gulick, A.M. | | Deposit date: | 2008-04-21 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of a 140 degrees domain movement in the two-step reaction catalyzed by 4-chlorobenzoate:CoA ligase.
Biochemistry, 47, 2008
|
|
3CW8
 
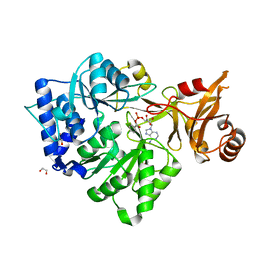 | | 4-Chlorobenzoyl-CoA Ligase/Synthetase, bound to 4CBA-Adenylate | | Descriptor: | 1,2-ETHANEDIOL, 4-chlorobenzoyl CoA ligase, 5'-O-[(S)-{[(4-chlorophenyl)carbonyl]oxy}(hydroxy)phosphoryl]adenosine | | Authors: | Reger, A.S, Cao, J, Wu, R, Dunaway-Mariano, D, Gulick, A.M. | | Deposit date: | 2008-04-21 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural characterization of a 140 degrees domain movement in the two-step reaction catalyzed by 4-chlorobenzoate:CoA ligase.
Biochemistry, 47, 2008
|
|
1SUQ
 
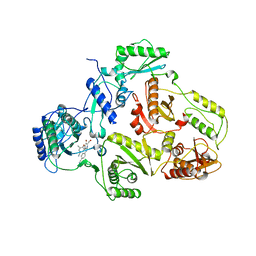 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R185545 | | Descriptor: | (6-[4-(AMINOMETHYL)-2,6-DIMETHYLPHENOXY]-2-{[4-(AMINOMETHYL)PHENYL]AMINO}-5-BROMOPYRIMIDIN-4-YL)METHANOL, MAGNESIUM ION, REVERSE TRANSCRIPTASE | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2004-03-26 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants
J.Med.Chem., 47, 2004
|
|
1SV5
 
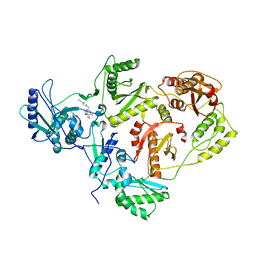 | | CRYSTAL STRUCTURE OF K103N MUTANT HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R165335 | | Descriptor: | 4-({6-AMINO-5-BROMO-2-[(4-CYANOPHENYL)AMINO]PYRIMIDIN-4-YL}OXY)-3,5-DIMETHYLBENZONITRILE, Reverse Transcriptase | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2004-03-27 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants
J.Med.Chem., 47, 2004
|
|
1S6P
 
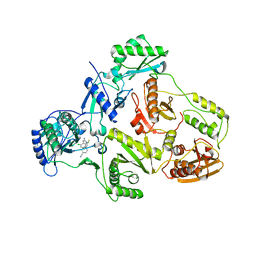 | | CRYSTAL STRUCTURE OF HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R100943 | | Descriptor: | 1-(4-CYANO-PHENYL)-3-[2-(2,6-DICHLORO-PHENYL)-1-IMINO-ETHYL]-THIOUREA, MAGNESIUM ION, POL polyprotein [Contains: Reverse transcriptase] | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2004-01-26 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants
J.Med.Chem., 47, 2004
|
|
1S9E
 
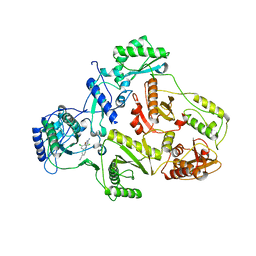 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R129385 | | Descriptor: | 4-[4-AMINO-6-(2,6-DICHLORO-PHENOXY)-[1,3,5]TRIAZIN-2-YLAMINO]-BENZONITRILE, POL polyprotein [Contains: Reverse transcriptase], POL polyprotein [Contains:Reverse transcriptase] | | Authors: | Das, K, Clark Jr, A.D, Ludovici, D.W, Kukla, M.J, Decorte, B, Lewi, P.J, Hughes, S.H, Janssen, P.A, Arnold, E. | | Deposit date: | 2004-02-04 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants.
J.Med.Chem., 47, 2004
|
|
1S9G
 
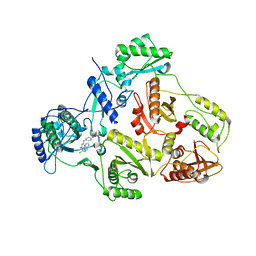 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R120394. | | Descriptor: | 4-[4-AMINO-6-(5-CHLORO-1H-INDOL-4-YLMETHYL)-[1,3,5]TRIAZIN-2-YLAMINO]-BENZONITRILE, POL polyprotein [Contains: Reverse transcriptase] | | Authors: | Das, K, Clark Jr, A.D, Ludovici, D.W, Kukla, M.J, Decorte, B, Lewi, P.J, Hughes, S.H, Janssen, P.A, Arnold, E. | | Deposit date: | 2004-02-04 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants.
J.Med.Chem., 47, 2004
|
|
1S6Q
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R147681 | | Descriptor: | 4-[4-(2,4,6-TRIMETHYL-PHENYLAMINO)-PYRIMIDIN-2-YLAMINO]-BENZONITRILE, POL polyprotein [Contains: Reverse transcriptase] | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2004-01-26 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants.
J.Med.Chem., 47, 2004
|
|
8GV5
 
 | | Crystal structure of PN-SIA28 in complex with influenza hemagglutinin A/swine/Guangdong/104/2013 (H1N1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain, ... | | Authors: | Chen, Y, Song, H, Qi, J, Gao, G.F. | | Deposit date: | 2022-09-14 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for a human broadly neutralizing influenza A hemagglutinin stem-specific antibody including H17/18 subtypes.
Nat Commun, 13, 2022
|
|
8GV6
 
 | | Crystal structure of PN-SIA28 in complex with influenza hemagglutinin H14 (A/long-tailed duck/Wisconsin/10OS3912/2010) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin H14-HA1, ... | | Authors: | Chen, Y, Song, H, Qi, J, Gao, G.F. | | Deposit date: | 2022-09-14 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis for a human broadly neutralizing influenza A hemagglutinin stem-specific antibody including H17/18 subtypes.
Nat Commun, 13, 2022
|
|
8GV7
 
 | | Crystal structure of PN-SIA28 in complex with influenza hemagglutinin H18 A/flat-faced bat/Peru/033/2010 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin H18-HA1, ... | | Authors: | Chen, Y, Song, H, Qi, J, Gao, G.F. | | Deposit date: | 2022-09-14 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for a human broadly neutralizing influenza A hemagglutinin stem-specific antibody including H17/18 subtypes.
Nat Commun, 13, 2022
|
|
8GV4
 
 | | A human broadly neutralizing influenza A hemagglutinin stem-specific antibody PN-SIA28 | | Descriptor: | PN-SIA28 heavy chain, PN-SIA28 light chain | | Authors: | Chen, Y, Song, H, Qi, J, Gao, G.F. | | Deposit date: | 2022-09-14 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for a human broadly neutralizing influenza A hemagglutinin stem-specific antibody including H17/18 subtypes.
Nat Commun, 13, 2022
|
|
7XQ5
 
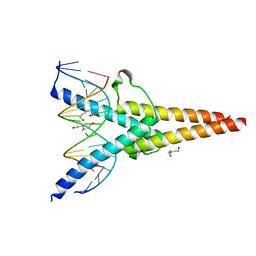 | | Crystal structure of ScIno2p-ScIno4p bound promoter DNA | | Descriptor: | DNA (5'-D(*GP*AP*TP*TP*TP*TP*CP*AP*CP*AP*TP*GP*CP*AP*G)-3'), DNA (5'-D(P*CP*CP*TP*GP*CP*AP*TP*GP*TP*GP*AP*AP*AP*AP*T)-3'), HEXANE-1,6-DIOL, ... | | Authors: | Khan, M.H, Lu, X. | | Deposit date: | 2022-05-06 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Analysis of Ino2p/Ino4p Mutual Interactions and Their Binding Interface with Promoter DNA.
Int J Mol Sci, 23, 2022
|
|
