7ZF7
 
 | | SARS-CoV-2 Omicron BA.2 RBD in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZR8
 
 | | OMI-38 FAB IN COMPLEX WITH SARS-COV-2 BETA SPIKE RBD (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Omi-38 Fab light chain, Omi-38 fab heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2022-05-03 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
2JJW
 
 | | Structure of human signal regulatory protein (sirp) gamma | | Descriptor: | SIGNAL REGULATORY PROTEIN GAMMA | | Authors: | Hatherley, D, Graham, S.C, Turner, J, Harlos, K, Stuart, D.I, Barclay, A.N. | | Deposit date: | 2008-04-22 | | Release date: | 2008-08-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Paired Receptor Specificity Explained by Structures of Signal Regulatory Proteins Alone and Complexed with Cd47.
Mol.Cell, 31, 2008
|
|
2JLF
 
 | | STRUCTURAL EXPLANATION FOR THE ROLE OF MN IN THE ACTIVITY OF PHI6 RNA- DEPENDENT RNA POLYMERASE | | Descriptor: | MANGANESE (II) ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Poranen, M.M, Salgado, P.S, Koivunen, M.R.L, Wright, S, Bamford, D.H, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2008-09-08 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Explanation for the Role of Mn2+ in the Activity of {Phi}6 RNA-Dependent RNA Polymerase.
Nucleic Acids Res., 36, 2008
|
|
2JLG
 
 | | STRUCTURAL EXPLANATION FOR THE ROLE OF MN IN THE ACTIVITY OF PHI6 RNA-DEPENDENT RNA POLYMERASE | | Descriptor: | 5'-D(*DT DT DT DC DCP)-3', GUANOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Poranen, M.M, Salgado, P.S, Koivunen, M.R.L, Wright, S, Bamford, D.H, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2008-09-09 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Explanation for the Role of Mn2+ in the Activity of {Phi}6 RNA-Dependent RNA Polymerase.
Nucleic Acids Res., 36, 2008
|
|
2JJX
 
 | | THE CRYSTAL STRUCTURE OF UMP KINASE FROM BACILLUS ANTHRACIS (BA1797) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, URIDYLATE KINASE | | Authors: | Meier, C, Carter, L.G, Mancini, E.J, Owens, R.J, Stuart, D.I, Esnouf, R.M, Oxford Protein Production Facility (OPPF), Structural Proteomics in Europe (SPINE) | | Deposit date: | 2008-04-23 | | Release date: | 2008-07-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | The Crystal Structure of Ump Kinase from Bacillus Anthracis (Ba1797) Reveals an Allosteric Nucleotide-Binding Site.
J.Mol.Biol., 381, 2008
|
|
7ZFA
 
 | | SARS-CoV-2 Omicron RBD in complex with Omi-6 and COVOX-150 Fabs | | Descriptor: | COVOX-150 heavy chain, COVOX-150 light chain, Omi-6 heavy chain, ... | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (4.24 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
2JJT
 
 | | Structure of human CD47 in complex with human signal regulatory protein (SIRP) alpha | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LEUKOCYTE SURFACE ANTIGEN CD47, TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE SUBSTRATE 1 | | Authors: | Hatherley, D, Graham, S.C, Turner, J, Harlos, K, Stuart, D.I, Barclay, A.N. | | Deposit date: | 2008-04-22 | | Release date: | 2008-08-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Paired receptor specificity explained by structures of signal regulatory proteins alone and complexed with CD47.
Mol. Cell, 31, 2008
|
|
7ZXU
 
 | | SARS-CoV-2 Omicron BA.4/5 RBD in complex with Beta-27 Fab and C1 nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-27 heavy chain, Beta-27 light chain, ... | | Authors: | Huo, J, Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2022-05-23 | | Release date: | 2022-06-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Antibody escape of SARS-CoV-2 Omicron BA.4 and BA.5 from vaccine and BA.1 serum.
Cell, 185, 2022
|
|
2JHP
 
 | | The structure of bluetongue virus VP4 reveals a multifunctional RNA- capping production-line | | Descriptor: | GUANINE, S-ADENOSYL-L-HOMOCYSTEINE, VP4 CORE PROTEIN | | Authors: | Sutton, G, Grimes, J.M, Stuart, D.I, Roy, P. | | Deposit date: | 2007-02-23 | | Release date: | 2007-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Bluetongue Virus Vp4 is an RNA-Capping Assembly Line.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2J7N
 
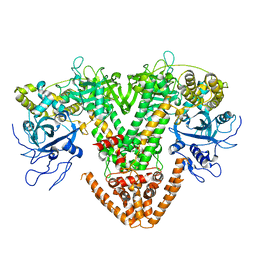 | | Structure of the RNAi polymerase from Neurospora crassa | | Descriptor: | GLYCEROL, MAGNESIUM ION, RNA-DEPENDENT RNA POLYMERASE | | Authors: | Salgado, P.S, Koivunen, M.R.L, Makeyev, E.V, Bamford, D.H, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2006-10-13 | | Release date: | 2006-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structure of an Rnai Polymerase Links RNA Silencing and Transcription.
Plos Biol., 4, 2006
|
|
2JH9
 
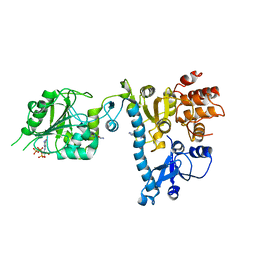 | | The structure of bluetongue virus VP4 reveals a multifunctional RNA- capping production-line | | Descriptor: | GUANINE, GUANOSINE-5'-TRIPHOSPHATE, VP4 CORE PROTEIN | | Authors: | Sutton, G, Grimes, J.M, Stuart, D.I, Roy, P. | | Deposit date: | 2007-02-21 | | Release date: | 2007-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Bluetongue Virus Vp4 is an RNA-Capping Assembly Line.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2JJV
 
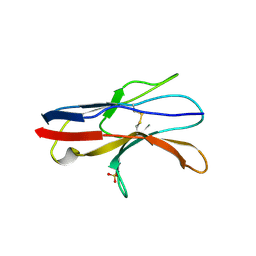 | | Structure of human signal regulatory protein (sirp) beta(2) | | Descriptor: | CHLORIDE ION, SIGNAL-REGULATORY PROTEIN BETA 1., SULFATE ION | | Authors: | Hatherley, D, Graham, S.C, Turner, J, Harlos, K, Stuart, D.I, Barclay, A.N. | | Deposit date: | 2008-04-22 | | Release date: | 2008-08-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Paired Receptor Specificity Explained by Structures of Signal Regulatory Proteins Alone and Complexed with Cd47.
Mol.Cell, 31, 2008
|
|
2W2S
 
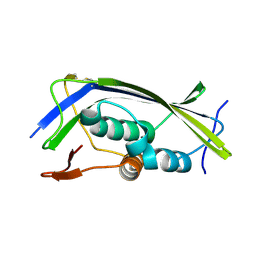 | | Structure of the Lagos bat virus matrix protein | | Descriptor: | MATRIX PROTEIN | | Authors: | Graham, S.C, Assenberg, R, Delmas, O, Verma, A, Gholami, A, Talbi, C, Owens, R.J, Stuart, D.I, Grimes, J.M, Bourhy, H. | | Deposit date: | 2008-11-03 | | Release date: | 2009-01-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Rhabdovirus Matrix Protein Structures Reveal a Novel Mode of Self-Association.
Plos Pathog., 4, 2008
|
|
2JHC
 
 | | The structure of bluetongue virus VP4 reveals a multifunctional RNA- capping production-line | | Descriptor: | GUANINE, VP4 CORE PROTEIN | | Authors: | Sutton, G, Grimes, J.M, Stuart, D.I, Roy, P. | | Deposit date: | 2007-02-21 | | Release date: | 2007-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Bluetongue Virus Vp4 is an RNA-Capping Assembly Line.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2JL9
 
 | | Structural explanation for the role of Mn in the activity of phi6 RNA- dependent RNA polymerase | | Descriptor: | RNA-DIRECTED RNA POLYMERASE | | Authors: | Poranen, M.M, Salgado, P.S, Koivunen, M.R.L, Wright, S, Bamford, D.H, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2008-09-05 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Explanation for the Role of Mn2+ in the Activity of {Phi}6 RNA-Dependent RNA Polymerase.
Nucleic Acids Res., 36, 2008
|
|
2JHA
 
 | | The structure of bluetongue virus VP4 reveals a multifunctional RNA- capping production-line | | Descriptor: | DIGUANOSINE-5'-TRIPHOSPHATE, GUANINE, VP4 CORE PROTEIN | | Authors: | Sutton, G, Grimes, J.M, Stuart, D.I, Roy, P. | | Deposit date: | 2007-02-21 | | Release date: | 2007-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Bluetongue Virus Vp4 is an RNA-Capping Assembly Line.
Nat.Struct.Mol.Biol., 14, 2007
|
|
8A8N
 
 | |
2X7L
 
 | | Implications of the HIV-1 Rev dimer structure at 3.2A resolution for multimeric binding to the Rev response element | | Descriptor: | FAB HEAVY CHAIN, FAB LIGHT CHAIN, PROTEIN REV | | Authors: | DiMattia, M.A, Watts, N.R, Stahl, S.J, Rader, C, Wingfield, P.T, Stuart, D.I, Steven, A.C, Grimes, J.M. | | Deposit date: | 2010-03-01 | | Release date: | 2010-03-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Implications of the HIV-1 Rev Dimer Structure at 3. 2 A Resolution for Multimeric Binding to the Rev Response Element.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2JH8
 
 | | The structure of bluetongue virus VP4 reveals a multifunctional RNA- capping production-line | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, GUANINE, VP4 CORE PROTEIN | | Authors: | Sutton, G, Grimes, J.M, Stuart, D.I, Roy, P. | | Deposit date: | 2007-02-21 | | Release date: | 2007-04-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Bluetongue Virus Vp4 is an RNA-Capping Assembly Line.
Nat.Struct.Mol.Biol., 14, 2007
|
|
1LMW
 
 | | LMW U-PA Structure complexed with EGRCMK (GLU-GLY-ARG Chloromethyl Ketone) | | Descriptor: | L-alpha-glutamyl-N-{(1S)-4-{[amino(iminio)methyl]amino}-1-[(1S)-2-chloro-1-hydroxyethyl]butyl}glycinamide, UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | Authors: | Spraggon, G.S, Phillips, C, Nowak, U.K, Ponting, C.P, Saunders, D, Dobson, C.M, Stuart, D.I, Jones, E.Y. | | Deposit date: | 1995-07-26 | | Release date: | 1996-01-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of the catalytic domain of human urokinase-type plasminogen activator.
Structure, 3, 1995
|
|
2JJS
 
 | | Structure of human CD47 in complex with human signal regulatory protein (SIRP) alpha | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IODIDE ION, LEUKOCYTE SURFACE ANTIGEN CD47, ... | | Authors: | Hatherley, D, Graham, S.C, Turner, J, Harlos, K, Stuart, D.I, Barclay, A.N. | | Deposit date: | 2008-04-22 | | Release date: | 2008-08-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Paired receptor specificity explained by structures of signal regulatory proteins alone and complexed with CD47.
Mol. Cell, 31, 2008
|
|
1LKI
 
 | | THE CRYSTAL STRUCTURE AND BIOLOGICAL FUNCTION OF LEUKEMIA INHIBITORY FACTOR: IMPLICATIONS FOR RECEPTOR BINDING | | Descriptor: | LEUKEMIA INHIBITORY FACTOR | | Authors: | Robinson, R.C, Grey, L.M, Staunton, D, Stuart, D.I, Heath, J.K, Jones, E.Y. | | Deposit date: | 1994-12-12 | | Release date: | 1995-03-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure and biological function of leukemia inhibitory factor: implications for receptor binding.
Cell(Cambridge,Mass.), 77, 1994
|
|
1OLZ
 
 | | The ligand-binding face of the semaphorins revealed by the high resolution crystal structure of SEMA4D | | Descriptor: | SEMAPHORIN 4D | | Authors: | Love, C.A, Harlos, K, Mavaddat, N, Davis, S.J, Stuart, D.I, Jones, E.Y, Esnouf, R.M. | | Deposit date: | 2003-08-19 | | Release date: | 2003-09-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Ligand-Binding Face of the Semaphorins Revealed by the High-Resolution Crystal Structure of Sema4D
Nat.Struct.Biol., 10, 2003
|
|
1OOP
 
 | | The Crystal Structure of Swine Vesicular Disease Virus | | Descriptor: | Coat protein VP1, Coat protein VP2, Coat protein VP3, ... | | Authors: | Fry, E.E, Knowles, N.J, Newman, J.W.I, Wilsden, G, Rao, Z, King, A.M.Q, Stuart, D.I. | | Deposit date: | 2003-03-04 | | Release date: | 2003-04-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of Swine Vesicular Disease Virus and Implications for Host Adaptation
J.Virol., 77, 2003
|
|
