2NNN
 
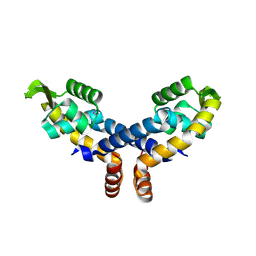 | | Crystal structure of probable transcriptional regulator from Pseudomonas aeruginosa | | Descriptor: | Probable transcriptional regulator | | Authors: | Chang, C, Evdokimova, E, Altamentova, S, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-10-24 | | Release date: | 2006-11-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of probable transcriptional regulator from Pseudomonas aeruginosa
To be Published
|
|
2NS0
 
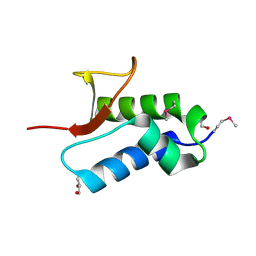 | | Crystal structure of protein RHA04536 from Rhodococcus sp | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Hypothetical protein | | Authors: | Chang, C, Skarina, T, Onopriyenko, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-11-02 | | Release date: | 2006-12-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Crystal structure of protein RHA04536 from Rhodococcus sp
To be Published
|
|
6MGL
 
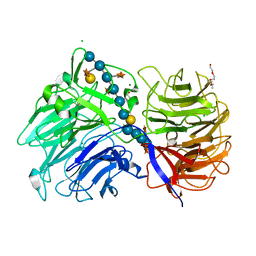 | | Crystal structure of the catalytic domain from GH74 enzyme PoGH74 from Paenibacillus odorifer, D60A mutant in complex with XXLG and XGXXLG xyloglucan | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Skarina, T, Arnal, G, Watanabe, N, Brumer, H, Savchenko, A. | | Deposit date: | 2018-09-14 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural enzymology reveals the molecular basis of substrate regiospecificity and processivity of an exemplar bacterial glycoside hydrolase family 74endo-xyloglucanase.
Biochem. J., 475, 2018
|
|
6MGJ
 
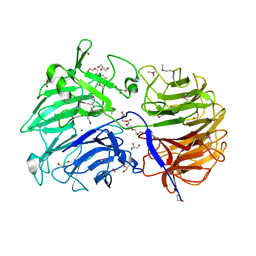 | | Crystal structure of the catalytic domain from GH74 enzyme PoGH74 from Paenibacillus odorifer, apoenzyme | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Skarina, T, Nocek, B, Arnal, G, Brumer, H, Savchenko, A. | | Deposit date: | 2018-09-14 | | Release date: | 2019-01-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural enzymology reveals the molecular basis of substrate regiospecificity and processivity of an exemplar bacterial glycoside hydrolase family 74endo-xyloglucanase.
Biochem. J., 475, 2018
|
|
6MGK
 
 | | Crystal structure of the catalytic domain from GH74 enzyme PoGH74 from Paenibacillus odorifer, in complex with XLX xyloglucan | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Skarina, T, Nocek, B, Arnal, G, Brumer, H, Savchenko, A. | | Deposit date: | 2018-09-14 | | Release date: | 2019-01-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural enzymology reveals the molecular basis of substrate regiospecificity and processivity of an exemplar bacterial glycoside hydrolase family 74endo-xyloglucanase.
Biochem. J., 475, 2018
|
|
1LXN
 
 | | X-RAY STRUCTURE OF MTH1187 NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET TT272 | | Descriptor: | HYPOTHETICAL PROTEIN MTH1187, SULFATE ION | | Authors: | Tao, X, Khayat, R, Christendat, D, Savchenko, A, Xu, X, Edwards, A, Arrowsmith, C.H, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-06-05 | | Release date: | 2003-07-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of MTH1187 and its Yeast Ortholog YBL001C
Proteins, 52, 2003
|
|
1LXJ
 
 | | X-RAY STRUCTURE OF YBL001c NORTHEAST STRUCTURAL GENOMICS (NESG) CONSORTIUM TARGET YTYst72 | | Descriptor: | HYPOTHETICAL 11.5KDA PROTEIN IN HTB2-NTH2 INTERGENIC REGION, SULFATE ION | | Authors: | Tao, X, Khayat, R, Christendat, D, Savchenko, A, Xu, X, Edwards, A, Arrowsmith, C.H, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-06-05 | | Release date: | 2003-07-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CRYSTAL STRUCTURES OF MTH1187 AND ITS YEAST ORTHOLOG YBL001C
Proteins, 52, 2003
|
|
3V77
 
 | | Crystal structure of a putative fumarylacetoacetate isomerase/hydrolase from Oleispira antarctica | | Descriptor: | ACETATE ION, D(-)-TARTARIC ACID, Putative fumarylacetoacetate isomerase/hydrolase, ... | | Authors: | Stogios, P.J, Kagan, O, Di Leo, R, Bochkarev, A, Edwards, A.M, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-12-20 | | Release date: | 2012-01-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
5VGC
 
 | | Crystal structure of the NleG5-1 effector (C200A) from Escherichia coli O157:H7 str. Sakai | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Borek, D, Valleau, D, Skarina, T, Jobin, M.C, Wawrzak, Z, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-04-10 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the NleG5-1 effector (C200A) from Escherichia coli O157:H7 str. Sakai
To Be Published
|
|
1NXH
 
 | | X-RAY STRUCTURE: NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET TT87 | | Descriptor: | MTH396 protein | | Authors: | Khayat, R, Savchenko, A, Edwards, A, Arowsmith, C, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-02-10 | | Release date: | 2004-02-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-RAY STRUCTURE OF MTH396
To be Published
|
|
1NN4
 
 | | Structural Genomics, RpiB/AlsB | | Descriptor: | Ribose 5-phosphate isomerase B | | Authors: | Zhang, R.G, Andersson, C.E, Mowbray, S.L, Savchenko, A, Skarina, T, Evdokimova, E, Beasley, S.L, Arrowsmith, C, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-01-12 | | Release date: | 2003-07-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The 2.2 A resolution structure of RpiB/AlsB from Escherichia coli illustrates a new approach to the ribose-5-phosphate isomerase reaction.
J.Mol.Biol., 332, 2003
|
|
1O8B
 
 | | Structure of Escherichia coli ribose-5-phosphate isomerase, RpiA, complexed with arabinose-5-phosphate. | | Descriptor: | 5-O-phosphono-beta-D-arabinofuranose, RIBOSE 5-PHOSPHATE ISOMERASE | | Authors: | Zhang, R.-g, Andersson, C.E, Savchenko, A, Skarina, T, Evdokimova, E, Beasley, S, Arrowsmith, C.H, Edwards, A.M, Joachimiak, A, Mowbray, S.L, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-11-26 | | Release date: | 2003-01-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structure of Escherichia Coli Ribose-5-Phosphate Isomerase: A Ubiquitous Enzyme of the Pentose Phosphate Pathway and the Calvin Cycle
Structure, 11, 2003
|
|
2RC3
 
 | | Crystal structure of CBS domain, NE2398 | | Descriptor: | BROMIDE ION, CBS domain, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Dong, A, Xu, X, Korniyenko, Y, Yakunin, A, Zheng, H, Walker, J.R, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-19 | | Release date: | 2007-10-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of CBS domain, NE2398.
To be Published
|
|
5T1P
 
 | | Crystal structure of the putative periplasmic solute-binding protein from Campylobacter jejuni | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ABC transporter, ... | | Authors: | Filippova, E.V, Wawrzsak, Z, Sandoval, J, Skarina, T, Grimshaw, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-08-19 | | Release date: | 2016-09-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the putative periplasmic solute-binding protein from Campylobacter jejuni
To Be Published
|
|
5U4O
 
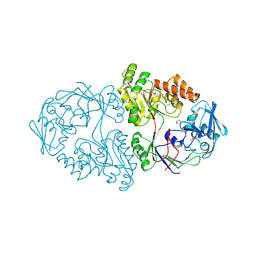 | | A 2.05A X-Ray Structureof A Bacterial Extracellular Solute-binding Protein, family 5 for Bacillus anthracis str. Ames | | Descriptor: | ABC transporter substrate-binding protein | | Authors: | Brunzelle, J.S, Wawrzak, Z, Sandoval, J, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-05 | | Release date: | 2017-03-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A 2.05A X-Ray Structureof A Bacterial Extracellular Solute-binding Protein, family 5 for Bacillus anthracis str. Ames
To Be Published
|
|
5TPM
 
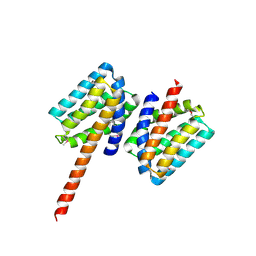 | | 2.8 Angstrom Crystal Structure of the C-terminal Dimerization Domain of Transcriptional Regulator PdhR from Escherichia coli. | | Descriptor: | Pyruvate dehydrogenase complex repressor | | Authors: | Minasov, G, Wawrzak, Z, Sandoval, J, Skarina, T, Grimshaw, S, Kwon, K, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-10-20 | | Release date: | 2016-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 2.8 Angstrom Crystal Structure of the C-terminal Dimerization Domain of Transcriptional Regulator PdhR from Escherichia coli.
To Be Published
|
|
5TPI
 
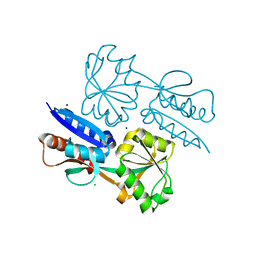 | | 1.47 Angstrom Crystal Structure of the C-terminal Substrate Binding Domain of LysR Family Transcriptional Regulator from Klebsiella pneumoniae. | | Descriptor: | CHLORIDE ION, Putative transcriptional regulator (LysR family), SODIUM ION | | Authors: | Minasov, G, Wawrzak, Z, Sandoval, J, Evdokimova, E, Grimshaw, S, Kwon, K, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-10-20 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | 1.47 Angstrom Crystal Structure of the C-terminal Substrate Binding Domain of LysR Family Transcriptional Regulator from Klebsiella pneumoniae.
To Be Published
|
|
2PZ9
 
 | | Crystal structure of putative transcriptional regulator SCO4942 from Streptomyces coelicolor | | Descriptor: | Putative regulatory protein, SULFATE ION | | Authors: | Filippova, E.V, Chruszcz, M, Xu, X, Zheng, H, Cymborowski, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-05-17 | | Release date: | 2007-06-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | In situ proteolysis for protein crystallization and structure determination.
Nat.Methods, 4, 2007
|
|
2P06
 
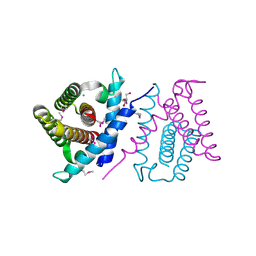 | | Crystal structure of a predicted coding region AF_0060 from Archaeoglobus fulgidus DSM 4304 | | Descriptor: | GLYCEROL, Hypothetical protein AF_0060, MAGNESIUM ION | | Authors: | Nocek, B, Xu, X, Koniyenko, Y, Yakounine, A, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-02-28 | | Release date: | 2007-03-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a predicted coding region AF_0060 from Archaeoglobus fulgidus DSM 4304
To be Published
|
|
3KQF
 
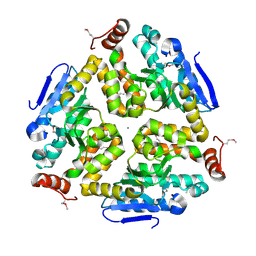 | | 1.8 Angstrom Resolution Crystal Structure of Enoyl-CoA Hydratase from Bacillus anthracis. | | Descriptor: | CALCIUM ION, CHLORIDE ION, Enoyl-CoA hydratase/isomerase family protein | | Authors: | Minasov, G, Halavaty, A, Wawrzak, Z, Skarina, T, Onopriyenko, O, Papazisi, L, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-11-17 | | Release date: | 2009-11-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 Angstrom Resolution Crystal Structure of Enoyl-CoA Hydratase from Bacillus anthracis.
TO BE PUBLISHED
|
|
3OMB
 
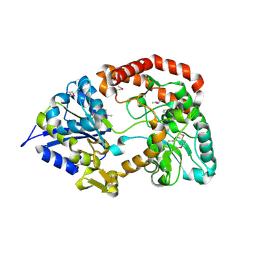 | | Crystal structure of extracellular solute-binding protein from Bifidobacterium longum subsp. infantis | | Descriptor: | Extracellular solute-binding protein, family 1, MAGNESIUM ION | | Authors: | Chang, C, Xu, X, Chin, S, Cui, H, Dong, A, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-26 | | Release date: | 2010-09-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of extracellular solute-binding protein from Bifidobacterium longum subsp. infantis
To be Published
|
|
3KUX
 
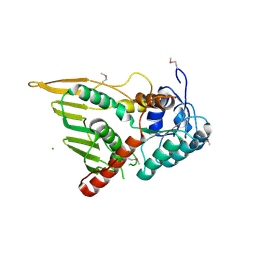 | | Structure of the YPO2259 putative oxidoreductase from Yersinia pestis | | Descriptor: | CHLORIDE ION, Putative oxidoreductase | | Authors: | Anderson, S.M, Wawrzak, Z, Gordon, E, Kwon, K, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-11-28 | | Release date: | 2009-12-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of the YPO2259 putative oxidoreductase from Yersinia pestis
To be Published
|
|
3KUU
 
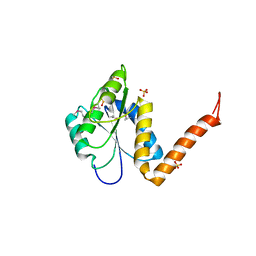 | | Structure of the PurE Phosphoribosylaminoimidazole Carboxylase Catalytic Subunit from Yersinia pestis | | Descriptor: | Phosphoribosylaminoimidazole carboxylase catalytic subunit PurE, SULFATE ION | | Authors: | Anderson, S.M, Wawrzak, Z, Brunzelle, J.S, Onopriyenko, O, Kwon, K, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-11-27 | | Release date: | 2009-12-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structure of the PurE Phosphoribosylaminoimidazole Carboxylase Catalytic Subunit from Yersinia pestis
To be Published
|
|
3LU2
 
 | | Structure of lmo2462, a Listeria monocytogenes amidohydrolase family putative dipeptidase | | Descriptor: | Lmo2462 protein, ZINC ION | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Hasseman, J, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-02-16 | | Release date: | 2010-03-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of lmo2462, a Listeria monocytogenes amidohydrolase family putative dipeptidase
To be Published
|
|
6BRD
 
 | | Crystal structure of rifampin monooxygenase from Streptomyces venezuelae, complexed with rifampin and FAD | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Cox, G, Kelso, J, Stogios, P.J, Savchenko, A, Anderson, W.F, Wright, G.D, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-11-30 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Rox, a Rifamycin Resistance Enzyme with an Unprecedented Mechanism of Action.
Cell Chem Biol, 25, 2018
|
|
