4FQM
 
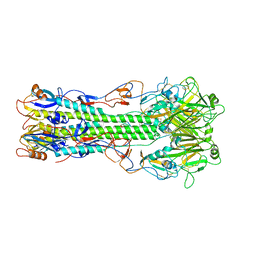 | | Structure of B/Brisbane/60/2008 Influenza Hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | Authors: | Dreyfus, C, Laursen, N.S, Wilson, I.A. | | Deposit date: | 2012-06-25 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Highly conserved protective epitopes on influenza B viruses.
Science, 337, 2012
|
|
4FQL
 
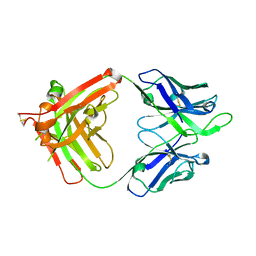 | | Influenza B HA Antibody (Fab) CR8033 | | Descriptor: | monoclonal antibody CR8033 heavy chain, monoclonal antibody CR8033 light chain | | Authors: | Dreyfus, C, Laursen, N.S, Wilson, I.A. | | Deposit date: | 2012-06-25 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Highly conserved protective epitopes on influenza B viruses.
Science, 337, 2012
|
|
4FQV
 
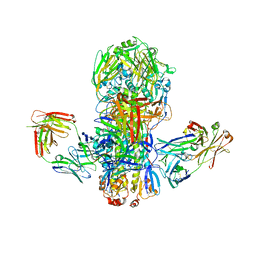 | | Crystal structure of broadly neutralizing antibody CR9114 bound to H7 influenza hemagglutinin | | Descriptor: | Antibody CR9114 heavy chain, Antibody CR9114 light chain, Hemagglutinin HA1, ... | | Authors: | Ekiert, D.C, Dreyfus, C, Wilson, I.A. | | Deposit date: | 2012-06-25 | | Release date: | 2012-08-22 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (5.75 Å) | | Cite: | Highly conserved protective epitopes on influenza B viruses.
Science, 337, 2012
|
|
4FNK
 
 | |
4FQK
 
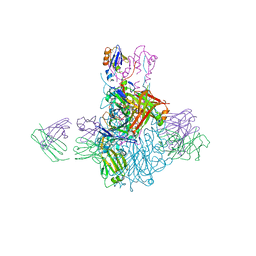 | | Influenza B/Brisbane/60/2008 hemagglutinin Fab CR8059 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody CR8059 Heavy Chain, ... | | Authors: | Dreyfus, C, Laursen, N.S, Wilson, I.A. | | Deposit date: | 2012-06-25 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (5.65 Å) | | Cite: | Highly conserved protective epitopes on influenza B viruses.
Science, 337, 2012
|
|
6K8H
 
 | | Crystal structure of an omega-transaminase from Sphaerobacter thermophilus | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, Aminotransferase class-III | | Authors: | Park, H.H, Kwon, S. | | Deposit date: | 2019-06-12 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the enzyme specificity of a novel omega-transaminase from the thermophilic bacterium Sphaerobacter thermophilus.
J.Struct.Biol., 208, 2019
|
|
4HB9
 
 | |
4FQH
 
 | | Crystal Structure of Fab CR9114 | | Descriptor: | 1,2-ETHANEDIOL, NITRATE ION, antibody CR9114 heavy chain, ... | | Authors: | Dreyfus, C, Wilson, I.A. | | Deposit date: | 2012-06-25 | | Release date: | 2012-08-22 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Highly conserved protective epitopes on influenza B viruses.
Science, 337, 2012
|
|
4FP8
 
 | |
4FQJ
 
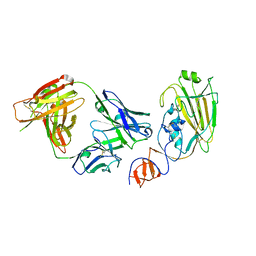 | | Influenza B/Florida/4/2006 hemagglutinin Fab CR8071 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, antibody CR8071 heavy chain, ... | | Authors: | Dreyfus, C, Laursen, N.S, Wilson, I.A. | | Deposit date: | 2012-06-25 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Highly conserved protective epitopes on influenza B viruses.
Science, 337, 2012
|
|
4FQY
 
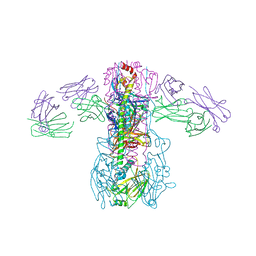 | | Crystal structure of broadly neutralizing antibody CR9114 bound to H3 influenza hemagglutinin | | Descriptor: | Antibody CR9114 heavy chain, Antibody CR9114 light chain, Hemagglutinin HA1, ... | | Authors: | Ekiert, D.C, Dreyfus, C, Wilson, I.A. | | Deposit date: | 2012-06-25 | | Release date: | 2012-08-22 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (5.253 Å) | | Cite: | Highly conserved protective epitopes on influenza B viruses.
Science, 337, 2012
|
|
4FQI
 
 | |
7YC5
 
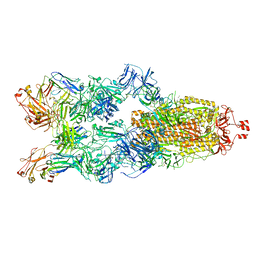 | | Cryo-EM structure of SARS-CoV-2 spike in complex with K202.B bispecific antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain from K202.B, bispecific antibody, ... | | Authors: | Yoo, Y, Cho, H.S. | | Deposit date: | 2022-06-30 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Novel bispecific human antibody platform specifically targeting a fully open spike conformation potently neutralizes multiple SARS-CoV-2 variants.
Antiviral Res., 212, 2023
|
|
3K1J
 
 | | Crystal structure of Lon protease from Thermococcus onnurineus NA1 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Cha, S.S, An, Y.J. | | Deposit date: | 2009-09-28 | | Release date: | 2010-09-22 | | Last modified: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Lon protease: molecular architecture of gated entry to a sequestered degradation chamber
Embo J., 29, 2010
|
|
3LDJ
 
 | | Crystal structure of aprotinin in complex with sucrose octasulfate: unusual interactions and implication for heparin binding | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, ACETATE ION, Pancreatic trypsin inhibitor | | Authors: | Yang, I.S, Kim, T.G, Park, B.S, Kim, K.H. | | Deposit date: | 2010-01-13 | | Release date: | 2010-09-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of aprotinin and its complex with sucrose octasulfate reveal multiple modes of interactions with implications for heparin binding.
Biochem.Biophys.Res.Commun., 2010
|
|
3LDM
 
 | |
4JY4
 
 | | Crystal structure of human Fab PGT121, a broadly reactive and potent HIV-1 neutralizing antibody | | Descriptor: | GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, PGT121 heavy chain, ... | | Authors: | Julien, J.-P, Diwanji, D.C, Burton, D.R, Wilson, I.A. | | Deposit date: | 2013-03-29 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Broadly neutralizing antibody PGT121 allosterically modulates CD4 binding via recognition of the HIV-1 gp120 V3 base and multiple surrounding glycans.
Plos Pathog., 9, 2013
|
|
4JY6
 
 | | Crystal structure of human Fab PGT123, a broadly reactive and potent HIV-1 neutralizing antibody | | Descriptor: | GLYCEROL, PGT123 heavy chain, PGT123 light chain, ... | | Authors: | Julien, J.-P, Diwanji, D.C, Burton, D.R, Wilson, I.A. | | Deposit date: | 2013-03-29 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Broadly Neutralizing Antibody PGT121 Allosterically Modulates CD4 Binding via Recognition of the HIV-1 gp120 V3 Base and Multiple Surrounding Glycans.
Plos Pathog., 9, 2013
|
|
3MYI
 
 | |
4TOY
 
 | |
4JM2
 
 | |
4JM4
 
 | | Crystal Structure of PGT 135 Fab | | Descriptor: | PGT 135 Heavy Chain, PGT 135 Light Chain | | Authors: | Kong, L, Wilson, I.A. | | Deposit date: | 2013-03-13 | | Release date: | 2013-05-29 | | Last modified: | 2013-07-17 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Supersite of immune vulnerability on the glycosylated face of HIV-1 envelope glycoprotein gp120.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4UC4
 
 | |
4JY5
 
 | | Crystal structure of human Fab PGT122, a broadly reactive and potent HIV-1 neutralizing antibody | | Descriptor: | GLYCEROL, PGT122 heavy chain, PGT122 light chain | | Authors: | Julien, J.-P, Diwanji, D.C, Burton, D.R, Wilson, I.A. | | Deposit date: | 2013-03-29 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Broadly neutralizing antibody PGT121 allosterically modulates CD4 binding via recognition of the HIV-1 gp120 V3 base and multiple surrounding glycans.
Plos Pathog., 9, 2013
|
|
1ZKJ
 
 | | Structural Basis for the Extended Substrate Spectrum of CMY-10, a Plasmid-Encoded Class C beta-lactamase | | Descriptor: | ACETIC ACID, ZINC ION, extended-spectrum beta-lactamase | | Authors: | Cha, S.S, Jung, H.I, An, Y.J, Lee, S.H. | | Deposit date: | 2005-05-03 | | Release date: | 2006-04-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for the extended substrate spectrum of CMY-10, a plasmid-encoded class C beta-lactamase.
Mol.Microbiol., 60, 2006
|
|
