8B6V
 
 | | Mp2Ba1 pre-pore | | Descriptor: | MAGNESIUM ION, Mpf2Ba1 | | Authors: | Marini, G, Poland, B, Leininger, C, Lukoyanova, N, Spielbauer, D, Barry, J, Altier, D, Lum, A, Scolaro, E, Perez Ortega, C, Yalpani, N, Sandahl, G, Mabry, T, Klever, J, Nowatzki, T, Zhao, J.Z, Sethi, A, Kassa, A, Crane, V, Lu, A, Nelson, M.E, Eswar, N, Topf, M, Saibil, H.R. | | Deposit date: | 2022-09-27 | | Release date: | 2023-05-31 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural journey of an insecticidal protein against western corn rootworm.
Nat Commun, 14, 2023
|
|
7O43
 
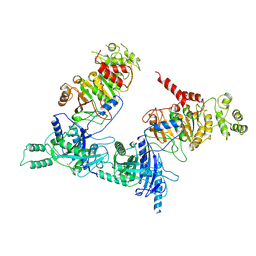 | |
7O3J
 
 | | O-layer structure (TrwH/VirB7, TrwF/VirB9CTD, TrwE/VirB10CTD) of the outer membrane core complex from the fully-assembled R388 type IV secretion system determined by cryo-EM. | | Descriptor: | TrwE protein, TrwF protein, TrwH protein | | Authors: | Mace, K, Vadakkepat, A.K, Lukoyanova, N, Waksman, G. | | Deposit date: | 2021-04-01 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Cryo-EM structure of a type IV secretion system.
Nature, 607, 2022
|
|
7O42
 
 | | TrwK/VirB4unbound trimer of dimers complex (with Hcp1) from the R388 type IV secretion system determined by cryo-EM. | | Descriptor: | TrwK protein,Protein hcp1 | | Authors: | Vadakkepat, A.K, Mace, K, Lukoyanova, N, Waksman, G. | | Deposit date: | 2021-04-04 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of a type IV secretion system.
Nature, 607, 2022
|
|
7O41
 
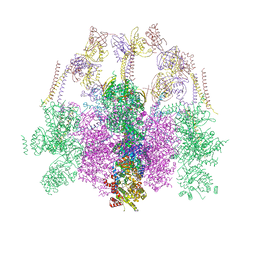 | | Hexameric composite model of the Inner Membrane Complex (IMC) with the Arches from the fully-assembled R388 type IV secretion system determined by cryo-EM. | | Descriptor: | TrwG protein, TrwK protein, TrwM protein | | Authors: | Mace, K, Vadakkepat, A.K, Lukoyanova, N, Waksman, G. | | Deposit date: | 2021-04-04 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Cryo-EM structure of a type IV secretion system.
Nature, 607, 2022
|
|
7O3V
 
 | | Stalk complex structure (TrwJ/VirB5-TrwI/VirB6) from the fully-assembled R388 type IV secretion system determined by cryo-EM. | | Descriptor: | TrwI protein, TrwJ protein | | Authors: | Mace, K, Vadakkepat, A.K, Lukoyanova, N, Waksman, G. | | Deposit date: | 2021-04-03 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of a type IV secretion system.
Nature, 607, 2022
|
|
7O3T
 
 | | I-layer structure (TrwF/VirB9NTD, TrwE/VirB10NTD) of the outer membrane core complex from the fully-assembled R388 type IV secretion system determined by cryo-EM. | | Descriptor: | TrwE protein, TrwF protein | | Authors: | Mace, K, Vadakkepat, A.K, Lukoyanova, N, Waksman, G. | | Deposit date: | 2021-04-03 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of a type IV secretion system.
Nature, 607, 2022
|
|
7OIU
 
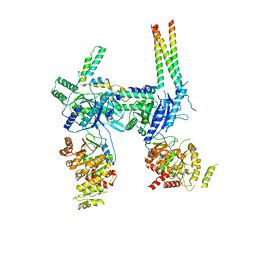 | | Inner Membrane Complex (IMC) protomer structure (TrwM/VirB3, TrwK/VirB4, TrwG/VirB8tails) from the fully-assembled R388 type IV secretion system determined by cryo-EM. | | Descriptor: | TrwG protein, TrwK protein, TrwM protein | | Authors: | Mace, K, Vadakkepat, A.K, Lukoyanova, N, Waksman, G. | | Deposit date: | 2021-05-12 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of a type IV secretion system.
Nature, 607, 2022
|
|
7NSU
 
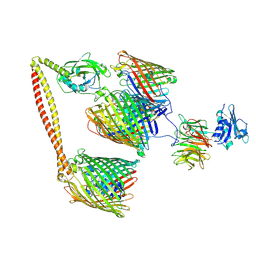 | | ColicinE9 intact translocation complex | | Descriptor: | Colicin-E9, Outer membrane protein F, Tol-Pal system protein TolB, ... | | Authors: | Webby, M.N, Kleanthous, C, Lukoyanova, N, Housden, N.G. | | Deposit date: | 2021-03-08 | | Release date: | 2021-08-11 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Porin threading drives receptor disengagement and establishes active colicin transport through Escherichia coli OmpF.
Embo J., 40, 2021
|
|
7NST
 
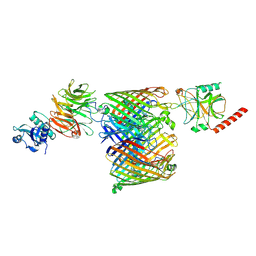 | | ColicinE9 partial translocation complex | | Descriptor: | Colicin-E9, Outer membrane protein F, Tol-Pal system protein TolB | | Authors: | Webby, M.N, Kleanthous, C, Lukoyanova, N, Housden, N.G. | | Deposit date: | 2021-03-08 | | Release date: | 2021-08-11 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Porin threading drives receptor disengagement and establishes active colicin transport through Escherichia coli OmpF.
Embo J., 40, 2021
|
|
7OK0
 
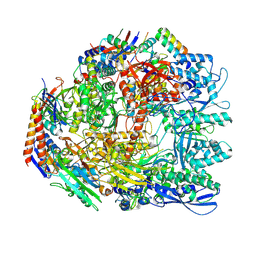 | | Cryo-EM structure of the Sulfolobus acidocaldarius RNA polymerase at 2.88 A | | Descriptor: | Conserved protein, DNA-directed RNA polymerase subunit A', DNA-directed RNA polymerase subunit A'', ... | | Authors: | Pilotto, S, Fouqueau, T, Lukoyanova, N, Sheppard, C, Lucas-Staat, S, Diaz-Santin, L.M, Matelska, D, Prangishvili, D, Cheung, A.C.M, Werner, F. | | Deposit date: | 2021-05-17 | | Release date: | 2021-08-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of RNA polymerase inhibition by viral and host factors.
Nat Commun, 12, 2021
|
|
7OQY
 
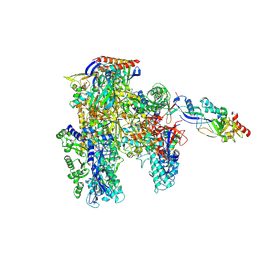 | | Cryo-EM structure of the cellular negative regulator TFS4 bound to the archaeal RNA polymerase | | Descriptor: | Conserved protein, DNA-directed RNA polymerase subunit A', DNA-directed RNA polymerase subunit A'', ... | | Authors: | Pilotto, S, Fouqueau, T, Lukoyanova, N, Sheppard, C, Lucas-Staat, S, Diaz-Santin, L.M, Matelska, D, Prangishvili, D, Cheung, A.C.M, Werner, F. | | Deposit date: | 2021-06-04 | | Release date: | 2021-08-25 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Structural basis of RNA polymerase inhibition by viral and host factors.
Nat Commun, 12, 2021
|
|
7OQ4
 
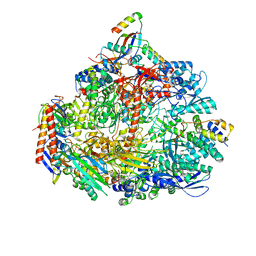 | | Cryo-EM structure of the ATV RNAP Inhibitory Protein (RIP) bound to the DNA-binding channel of the host's RNA polymerase | | Descriptor: | Conserved protein, DNA-directed RNA polymerase subunit A', DNA-directed RNA polymerase subunit A'', ... | | Authors: | Pilotto, S, Fouqueau, T, Lukoyanova, N, Sheppard, C, Lucas-Staat, S, Diaz-Santin, L.M, Matelska, D, Prangishvili, D, Cheung, A.C.M, Werner, F. | | Deposit date: | 2021-06-02 | | Release date: | 2021-08-25 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural basis of RNA polymerase inhibition by viral and host factors.
Nat Commun, 12, 2021
|
|
7OVB
 
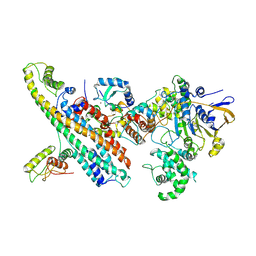 | | L. pneumophila Type IV Coupling Complex (T4CC) with density for DotY N-terminal and middle domains | | Descriptor: | DotY, DotZ, IcmJ (DotN), ... | | Authors: | Mace, K, Meir, A, Lukoyanova, N, Waksman, G. | | Deposit date: | 2021-06-14 | | Release date: | 2021-12-01 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Proteins DotY and DotZ modulate the dynamics and localization of the type IVB coupling complex of Legionella pneumophila.
Mol.Microbiol., 117, 2022
|
|
5OJS
 
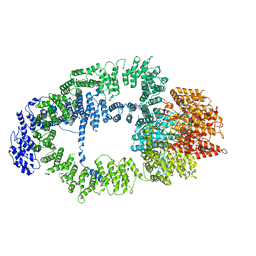 | |
6SX4
 
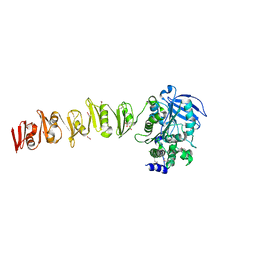 | |
6SWZ
 
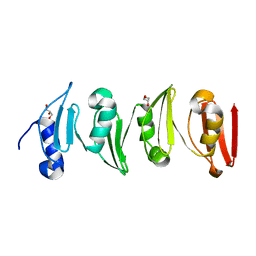 | |
7Q1V
 
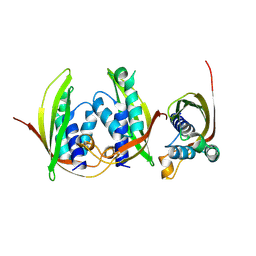 | |
6SZ9
 
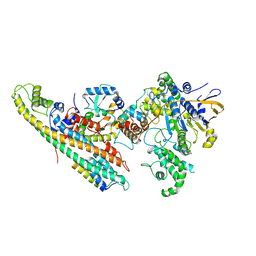 | | Type IV Coupling Complex (T4CC) from L. pneumophila. | | Descriptor: | DotY, DotZ, IcmJ (DotN), ... | | Authors: | Mace, K, Meir, A, Lukoyanova, N, Waksman, G. | | Deposit date: | 2019-10-02 | | Release date: | 2020-05-13 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanism of effector capture and delivery by the type IV secretion system from Legionella pneumophila.
Nat Commun, 11, 2020
|
|
5EX0
 
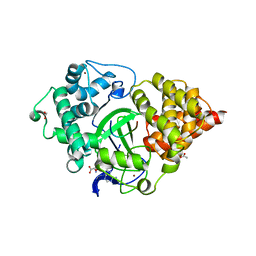 | | Crystal structure of human SMYD3 in complex with a MAP3K2 peptide | | Descriptor: | ACETIC ACID, Histone-lysine N-methyltransferase SMYD3, MAP3K2 peptide, ... | | Authors: | Fu, W, Liu, N, Qiao, Q, Wang, M, Min, J, Zhu, B, Xu, R.M, Yang, N. | | Deposit date: | 2015-11-23 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for Substrate Preference of SMYD3, a SET Domain-containing Protein Lysine Methyltransferase
J.Biol.Chem., 291, 2016
|
|
5EX3
 
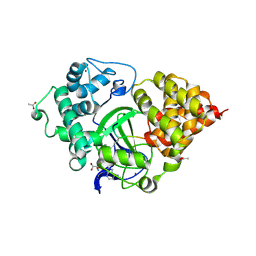 | | Crystal structure of human SMYD3 in complex with a VEGFR1 peptide | | Descriptor: | ACETIC ACID, Histone-lysine N-methyltransferase SMYD3, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Qiao, Q, Fu, W, Liu, N, Wang, M, Min, J, Zhu, B, Xu, R.M, Yang, N. | | Deposit date: | 2015-11-23 | | Release date: | 2016-03-09 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.408 Å) | | Cite: | Structural Basis for Substrate Preference of SMYD3, a SET Domain-containing Protein Lysine Methyltransferase
J.Biol.Chem., 291, 2016
|
|
2P4T
 
 | |
7QD8
 
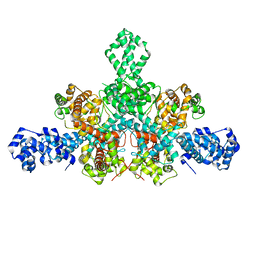 | | Cryo-EM structure of Tn4430 TnpA transposase from Tn3 family in apo state | | Descriptor: | Transposase for transposon Tn4430 | | Authors: | Shkumatov, A.V, Oger, C.A, Aryanpour, N, Hallet, B.F, Efremov, R.G. | | Deposit date: | 2021-11-26 | | Release date: | 2022-10-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insight into Tn3 family transposition mechanism.
Nat Commun, 13, 2022
|
|
7QD4
 
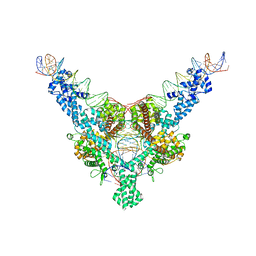 | | Cryo-EM structure of Tn4430 TnpA transposase from Tn3 family in complex with 100 bp long transposon end DNA | | Descriptor: | IR100 DNA substrate, none transferred strand, transferred strand, ... | | Authors: | Shkumatov, A.V, Oger, C.A, Aryanpour, N, Hallet, B.F, Efremov, R.G. | | Deposit date: | 2021-11-26 | | Release date: | 2022-10-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insight into Tn3 family transposition mechanism.
Nat Commun, 13, 2022
|
|
7QD6
 
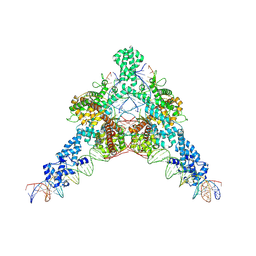 | | Cryo-EM structure of Tn4430 TnpA transposase from Tn3 family in complex with strand-transfer like DNA product | | Descriptor: | IR71st non transferred strand, IR71st transferred strand, Transposase for transposon Tn4430 | | Authors: | Shkumatov, A.V, Oger, C.A, Aryanpour, N, Hallet, B.F, Efremov, R.G. | | Deposit date: | 2021-11-26 | | Release date: | 2022-10-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insight into Tn3 family transposition mechanism.
Nat Commun, 13, 2022
|
|
