3H1X
 
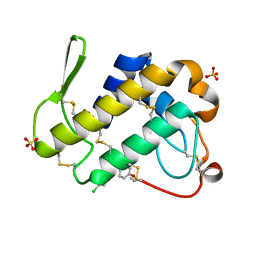 | | Simultaneous inhibition of anti-coagulation and inflammation: Crystal structure of phospholipase A2 complexed with indomethacin at 1.4 A resolution reveals the presence of the new common ligand binding site | | Descriptor: | INDOMETHACIN, Phospholipase A2 VRV-PL-VIIIa, SULFATE ION | | Authors: | Singh, N, Prem Kumar, R, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2009-04-14 | | Release date: | 2009-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Simultaneous inhibition of anti-coagulation and inflammation: crystal structure of phospholipase A2 complexed with indomethacin at 1.4 A resolution reveals the presence of the new common ligand-binding site
J.Mol.Recognit., 22, 2009
|
|
7F8R
 
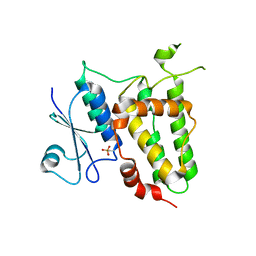 | |
6P9F
 
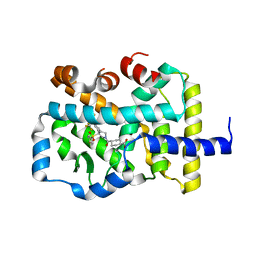 | | Crystal structure of RAR-related orphan receptor C (NHIS-RORGT(244-487)-L6-SRC1(678-692)) in complex with a phenyl (3-phenylpyrrolidin-3-yl)sulfone inhibitor | | Descriptor: | Nuclear receptor ROR-gamma, Nuclear receptor coactivator 1 chimera, trans-4-{(3R)-3-[(4-fluorophenyl)sulfonyl]-3-[4-(1,1,1,2,3,3,3-heptafluoropropan-2-yl)phenyl]pyrrolidine-1-carbonyl}cyclohexane-1-carboxylic acid | | Authors: | Sack, J. | | Deposit date: | 2019-06-10 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of potent, selective and orally bioavailable phenyl ((R)-3-phenylpyrrolidin-3-yl)sulfone analogues as ROR gamma t inverse agonists.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
1KBT
 
 | | SOLUTION STRUCTURE OF CARDIOTOXIN IV, NMR, 12 STRUCTURES | | Descriptor: | CTX IV | | Authors: | Jeng, J.Y, Kumar, T.K.S, Jayaraman, G, Yu, C. | | Deposit date: | 1996-07-22 | | Release date: | 1997-07-23 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Comparison of the hemolytic activity and solution structures of two snake venom cardiotoxin analogues which only differ in their N-terminal amino acid.
Biochemistry, 36, 1997
|
|
1KBS
 
 | | SOLUTION STRUCTURE OF CARDIOTOXIN IV, NMR, 1 STRUCTURE | | Descriptor: | CTX IV | | Authors: | Jeng, J.Y, Kumar, T.K.S, Jayaraman, G, Yu, C. | | Deposit date: | 1996-07-22 | | Release date: | 1997-07-23 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Comparison of the hemolytic activity and solution structures of two snake venom cardiotoxin analogues which only differ in their N-terminal amino acid.
Biochemistry, 36, 1997
|
|
5MHT
 
 | | TERNARY STRUCTURE OF HHAI METHYLTRANSFERASE WITH HEMIMETHYLATED DNA AND ADOHCY | | Descriptor: | DNA (5'-D(*CP*CP*AP*TP*GP*(5CM)P*GP*CP*TP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*AP*GP*CP*GP*CP*AP*TP*GP*G)-3'), PROTEIN (HHAI METHYLTRANSFERASE), ... | | Authors: | Cheng, X. | | Deposit date: | 1996-10-22 | | Release date: | 1997-07-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A structural basis for the preferential binding of hemimethylated DNA by HhaI DNA methyltransferase.
J.Mol.Biol., 263, 1996
|
|
1UBI
 
 | | SYNTHETIC STRUCTURAL AND BIOLOGICAL STUDIES OF THE UBIQUITIN SYSTEM. PART 1 | | Descriptor: | UBIQUITIN | | Authors: | Alexeev, D, Bury, S.M, Turner, M.A, Ogunjobi, O.M, Muir, T.W, Ramage, R, Sawyer, L. | | Deposit date: | 1994-02-03 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthetic, structural and biological studies of the ubiquitin system: the total chemical synthesis of ubiquitin.
Biochem.J., 299, 1994
|
|
1MHT
 
 | |
6LW8
 
 | | Structural basis for domain rotation during adenylation of active site K123 and fragment library screening against NAD+ -dependent DNA ligase from Mycobacterium tuberculosis | | Descriptor: | (4R)-4-(4-fluorophenyl)-4,5,6,7-tetrahydro-1H-imidazo[4,5-c]pyridine, DNA ligase A, GLYCEROL, ... | | Authors: | Ramachandran, R, Afsar, M, Shukla, A. | | Deposit date: | 2020-02-07 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structure based identification of first-in-class fragment inhibitors that target the NMN pocket of M. tuberculosis NAD + -dependent DNA ligase A.
J.Struct.Biol., 213, 2021
|
|
2SAS
 
 | |
4L69
 
 | |
8EUA
 
 | | Structure of SARS-CoV2 PLpro bound to a covalent inhibitor | | Descriptor: | Papain-like protease nsp3, SULFATE ION, ZINC ION, ... | | Authors: | Mathews, I.I, Pokhrel, S, Wakatsuki, S. | | Deposit date: | 2022-10-18 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Potent and selective covalent inhibition of the papain-like protease from SARS-CoV-2.
Nat Commun, 14, 2023
|
|
1VE3
 
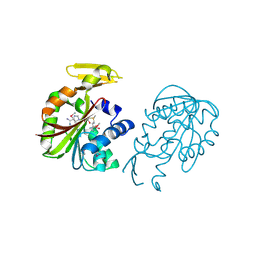 | | Crystal structure of PH0226 protein from Pyrococcus horikoshii OT3 | | Descriptor: | S-ADENOSYLMETHIONINE, hypothetical protein PH0226 | | Authors: | Lokanath, N.K, Yamamoto, H, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-26 | | Release date: | 2005-05-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of SAM-dependent methyltransferase from Pyrococcus horikoshii.
Acta Crystallogr.,Sect.F, 73, 2017
|
|
8A8N
 
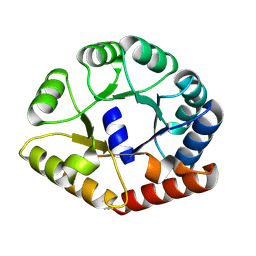 | |
8UDZ
 
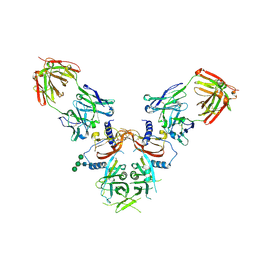 | | The Structure of LTBP-49247 Fab Bound to TGFbeta1 Small Latent Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LTBP-49247 Fab Heavy Chain, LTBP-49247 Fab Light Chain, ... | | Authors: | Streich Jr, F.C, Nicholls, S.B, Boston, C.J, Ramachandran, S. | | Deposit date: | 2023-09-29 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | An antibody that inhibits TGF-beta 1 release from latent extracellular matrix complexes attenuates the progression of renal fibrosis.
Sci.Signal., 17, 2024
|
|
7UVC
 
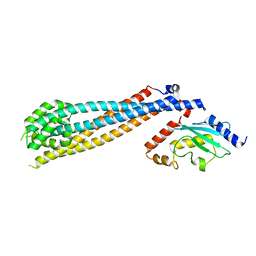 | | Rad6(P43L)-Bre1 Complex | | Descriptor: | E3 ubiquitin-protein ligase BRE1, Ubiquitin-conjugating enzyme E2 2 | | Authors: | Shukla, P.K, Chandrasekharan, M.B. | | Deposit date: | 2022-04-29 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structure and functional determinants of Rad6-Bre1 subunits in the histone H2B ubiquitin-conjugating complex.
Nucleic Acids Res., 51, 2023
|
|
7UV8
 
 | | Rad6-Bre1 Complex | | Descriptor: | E3 ubiquitin-protein ligase BRE1, Ubiquitin-conjugating enzyme E2 2 | | Authors: | Shukla, P.K, Chandrasekharan, M.B. | | Deposit date: | 2022-04-29 | | Release date: | 2023-01-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and functional determinants of Rad6-Bre1 subunits in the histone H2B ubiquitin-conjugating complex.
Nucleic Acids Res., 51, 2023
|
|
1R5N
 
 | | Crystal Structure Analysis of sup35 complexed with GDP | | Descriptor: | Eukaryotic peptide chain release factor GTP-binding subunit, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Kong, C, Song, H. | | Deposit date: | 2003-10-10 | | Release date: | 2004-05-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure and functional analysis of the eukaryotic class II release factor eRF3 from S. pombe
Mol.Cell, 14, 2004
|
|
1R5O
 
 | | crystal structure analysis of sup35 complexed with GMPPNP | | Descriptor: | Eukaryotic peptide chain release factor GTP-binding subunit, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Kong, C, Song, H. | | Deposit date: | 2003-10-11 | | Release date: | 2004-05-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure and functional analysis of the eukaryotic class II release factor eRF3 from S. pombe
Mol.Cell, 14, 2004
|
|
1R5B
 
 | | Crystal structure analysis of sup35 | | Descriptor: | Eukaryotic peptide chain release factor GTP-binding subunit | | Authors: | Kong, C, Song, H. | | Deposit date: | 2003-10-10 | | Release date: | 2004-05-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure and functional analysis of the eukaryotic class II release factor eRF3 from S. pombe
Mol.Cell, 14, 2004
|
|
4MHT
 
 | | TERNARY STRUCTURE OF HHAI METHYLTRANSFERASE WITH NATIVE DNA AND ADOHCY | | Descriptor: | DNA (5'-D(*GP*AP*TP*AP*GP*(5CM)P*GP*CP*TP*AP*TP*C)-3'), DNA (5'-D(*TP*GP*AP*TP*AP*GP*(5CM)P*GP*CP*TP*AP*TP*C)-3'), PROTEIN (HHAI METHYLTRANSFERASE (E.C.2.1.1.73)), ... | | Authors: | Cheng, X. | | Deposit date: | 1996-07-24 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Enzymatic C5-cytosine methylation of DNA: mechanistic implications of new crystal structures for HhaL methyltransferase-DNA-AdoHcy complexes.
J.Mol.Biol., 261, 1996
|
|
7U0Q
 
 | |
7U0X
 
 | |
6D2P
 
 | |
7UOW
 
 | | SARS-Cov2 S protein structure in complex with neutralizing monoclonal antibody 034_32 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Monoclonal antibody 034_32 heavy chain, ... | | Authors: | Patel, A, Ortlund, E. | | Deposit date: | 2022-04-14 | | Release date: | 2023-04-19 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Molecular basis of SARS-CoV-2 Omicron variant evasion from shared neutralizing antibody response.
Structure, 31, 2023
|
|
