5WWO
 
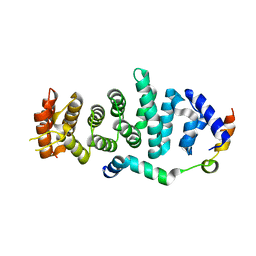 | | Crystal structure of Enp1 | | Descriptor: | Essential nuclear protein 1, Protein LTV1 | | Authors: | Ye, K, Zhang, W. | | Deposit date: | 2017-01-03 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular architecture of the 90S small subunit pre-ribosome
Elife, 6, 2017
|
|
7U0N
 
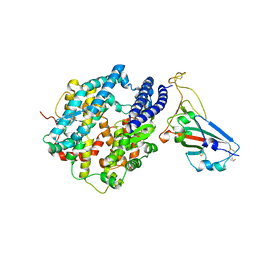 | | Crystal structure of chimeric omicron RBD (strain BA.1) complexed with human ACE2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Geng, Q, Shi, K, Ye, G, Zhang, W, Aihara, H, Li, F. | | Deposit date: | 2022-02-18 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural Basis for Human Receptor Recognition by SARS-CoV-2 Omicron Variant BA.1.
J.Virol., 96, 2022
|
|
5XKA
 
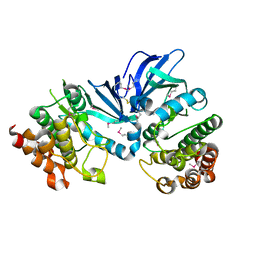 | | Crystal structure of M.tuberculosis PknI kinase domain | | Descriptor: | Serine/threonine-protein kinase PknI | | Authors: | Yan, Q, Jiang, D, Qian, L, Zhang, Q, Zhang, W, Zhou, W, Mi, K, Guddat, L, Yang, H, Rao, Z. | | Deposit date: | 2017-05-06 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Structural Insight into the Activation of PknI Kinase from M. tuberculosis via Dimerization of the Extracellular Sensor Domain.
Structure, 25, 2017
|
|
9KR5
 
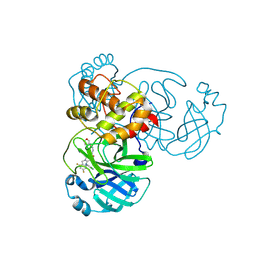 | | Crystal structure of SARS-CoV-2 main protease in complex with compound 3 | | Descriptor: | (6~{E})-6-(6-chloranyl-2-methyl-indazol-5-yl)imino-3-[5-[(3~{S})-oxolan-3-yl]oxypyridin-3-yl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazinane-2,4-dione, 3C-like proteinase nsp5 | | Authors: | Zhong, Y, Zhao, L, Zhang, W, Peng, W. | | Deposit date: | 2024-11-27 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Expanding the utilization of binding pockets proves to be effective for noncovalent small molecule inhibitors against SARS-CoV-2 M pro.
Eur.J.Med.Chem., 289, 2025
|
|
9KSK
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with compound 10 | | Descriptor: | 3C-like proteinase, 4-[4-chloranyl-2-[[(6E)-6-(6-chloranyl-2-methyl-indazol-5-yl)imino-3-(5-methylpyridin-3-yl)-2,4-bis(oxidanylidene)-1,3,5-triazinan-1-yl]methyl]-5-fluoranyl-phenoxy]-2-fluoranyl-benzenecarbonitrile | | Authors: | Zhong, Y, Zhao, L, Zhang, W, Peng, W. | | Deposit date: | 2024-11-29 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Expanding the utilization of binding pockets proves to be effective for noncovalent small molecule inhibitors against SARS-CoV-2 M pro.
Eur.J.Med.Chem., 289, 2025
|
|
9KSI
 
 | | Crystal Structure of SARS-CoV-2 main protease in complex with compound 5 | | Descriptor: | (6E)-1-[[5-chloranyl-4-fluoranyl-2-(4-fluoranylphenoxy)phenyl]methyl]-6-(6-chloranyl-2-methyl-indazol-5-yl)imino-3-(5-methoxypyridin-3-yl)-1,3,5-triazinane-2,4-dione, 3C-like proteinase nsp5 | | Authors: | Zhong, Y, Zhao, L, Zhang, W, Peng, W. | | Deposit date: | 2024-11-29 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Expanding the utilization of binding pockets proves to be effective for noncovalent small molecule inhibitors against SARS-CoV-2 M pro.
Eur.J.Med.Chem., 289, 2025
|
|
9KSH
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with compound 1 | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-pyridin-3-yl-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Zhong, Y, Zhao, L, Zhang, W, Peng, W. | | Deposit date: | 2024-11-29 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Expanding the utilization of binding pockets proves to be effective for noncovalent small molecule inhibitors against SARS-CoV-2 M pro.
Eur.J.Med.Chem., 289, 2025
|
|
9KSJ
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with compound 8 | | Descriptor: | 3-[[(6E)-6-(6-chloranyl-2-methyl-indazol-5-yl)imino-3-[5-(2-methoxyethoxy)pyridin-3-yl]-2,4-bis(oxidanylidene)-1,3,5-triazinan-1-yl]methyl]-4-methyl-benzenecarbonitrile, 3C-like proteinase nsp5 | | Authors: | Zhong, Y, Zhao, L, Zhang, W, Peng, W. | | Deposit date: | 2024-11-29 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Expanding the utilization of binding pockets proves to be effective for noncovalent small molecule inhibitors against SARS-CoV-2 M pro.
Eur.J.Med.Chem., 289, 2025
|
|
2L89
 
 | |
5K0K
 
 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC2434 | | Descriptor: | 15-{4-[(4-methylpiperazin-1-yl)methyl]phenyl}-4,5,6,7,9,10,11,12-octahydro-2,16-(azenometheno)pyrrolo[2,1-d][1,3,5,9]te traazacyclotetradecin-8(3H)-one, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Wang, X, Liu, J, Zhang, W, Stashko, M.A, Nichols, J, DeRyckere, D, Miley, M.J, Norris-Drouin, J, Chen, Z, Machius, M, Wood, E, Graham, D.K, Earp, H.S, Graham, K, Kireev, D, Frye, S.V. | | Deposit date: | 2016-05-17 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.545 Å) | | Cite: | Design and Synthesis of Novel Macrocyclic Mer Tyrosine Kinase Inhibitors.
ACS Med Chem Lett, 7, 2016
|
|
5K0X
 
 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC2541 | | Descriptor: | (7S)-7-amino-N-[(4-fluorophenyl)methyl]-8-oxo-2,9,16,18,21-pentaazabicyclo[15.3.1]henicosa-1(21),17,19-triene-20-carboxamide, CHLORIDE ION, Tyrosine-protein kinase Mer | | Authors: | McIver, A.L, Zhang, W, Liu, Q, Jiang, X, Stashko, M.A, Nichols, J, Miley, M.J, Norris-Drouin, J, Machius, M, DeRyckere, D, Wood, E, Graham, D.K, Earp, H.S, Kireev, D, Frye, S.V, Wang, X. | | Deposit date: | 2016-05-17 | | Release date: | 2017-02-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.231 Å) | | Cite: | Discovery of Macrocyclic Pyrimidines as MerTK-Specific Inhibitors.
ChemMedChem, 12, 2017
|
|
8TAR
 
 | | APC/C-CDH1-UBE2C-Ubiquitin-CyclinB-NTD | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Bodrug, T, Welsh, K.A, Bolhuis, D.L, Paulakonis, E, Martinez-Chacin, R.C, Liu, B, Pinkin, N, Bonacci, T, Cui, L, Xu, P, Roscow, O, Amann, S.J, Grishkovskaya, I, Emanuele, M.J, Harrison, J.S, Steimel, J.P, Hahn, K.M, Zhang, W, Zhong, E, Haselbach, D, Brown, N.G. | | Deposit date: | 2023-06-27 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Time-resolved cryo-EM (TR-EM) analysis of substrate polyubiquitination by the RING E3 anaphase-promoting complex/cyclosome (APC/C).
Nat.Struct.Mol.Biol., 30, 2023
|
|
8TAU
 
 | | APC/C-CDH1-UBE2C-UBE2S-Ubiquitin-CyclinB | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Bodrug, T, Welsh, K.A, Bolhuis, D.L, Paulakonis, E, Martinez-Chacin, R.C, Liu, B, Pinkin, N, Bonacci, T, Cui, L, Xu, P, Roscow, O, Amann, S.J, Grishkovskaya, I, Emanuele, M.J, Harrison, J.S, Steimel, J.P, Hahn, K.M, Zhang, W, Zhong, E, Haselbach, D, Brown, N.G. | | Deposit date: | 2023-06-27 | | Release date: | 2023-09-27 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Time-resolved cryo-EM (TR-EM) analysis of substrate polyubiquitination by the RING E3 anaphase-promoting complex/cyclosome (APC/C).
Nat.Struct.Mol.Biol., 30, 2023
|
|
9J56
 
 | | Functional Investigation of the SAM-Dependent Methyltransferases Rdmb in Anthracycline Biosynthesis | | Descriptor: | (7~{S},9~{S})-7-[(2~{R},4~{S},5~{S},6~{S})-4-azanyl-6-methyl-5-oxidanyl-oxan-2-yl]oxy-9-ethyl-4-methoxy-6,9,11-tris(oxidanyl)-8,10-dihydro-7~{H}-tetracene-5,12-dione, Aclacinomycin 10-hydroxylase RdmB, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Yang, Q.Y, Sang, M.L, Zhang, W. | | Deposit date: | 2024-08-11 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional investigation of the SAM-dependent methyltransferase RdmB in anthracycline biosynthesis.
Synth Syst Biotechnol, 10, 2025
|
|
9KQA
 
 | | Cryo-EM structure of human VMAT2 in complex with Tetrabenazine | | Descriptor: | (3S,5R,11bS)-9,10-dimethoxy-3-(2-methylpropyl)-1,3,4,6,7,11b-hexahydro-2H-pyrido[2,1-a]isoquinolin-2-one, Soluble cytochrome b562,Synaptic vesicular amine transporter | | Authors: | Wei, F, Zhang, W, Zhang, Y. | | Deposit date: | 2024-11-25 | | Release date: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Drug inhibition and substrate transport mechanisms of human VMAT2.
Nat Commun, 16, 2025
|
|
9KQE
 
 | |
9KQ8
 
 | |
5VKQ
 
 | | Structure of a mechanotransduction ion channel Drosophila NOMPC in nanodisc | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, No mechanoreceptor potential C isoform L | | Authors: | Jin, P, Bulkley, D, Guo, Y, Zhang, W, Guo, Z, Huynh, W, Wu, S, Meltzer, S, Chen, T, Jan, L.Y, Jan, Y.-N, Cheng, Y. | | Deposit date: | 2017-04-22 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Electron cryo-microscopy structure of the mechanotransduction channel NOMPC.
Nature, 547, 2017
|
|
8F24
 
 | | Mirror-image RNA octamer containing 2'-OMe-L-uridine | | Descriptor: | Mirror-image RNA 0G-XEC-0G-0U-0A-0C-0A-0C, SULFATE ION | | Authors: | Dantsu, Y, Zhang, W. | | Deposit date: | 2022-11-07 | | Release date: | 2023-04-12 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Derivatization of Mirror-Image l-Nucleic Acids with 2'-OMe Modification for Thermal and Structural Stabilization.
Chembiochem, 24, 2023
|
|
4YY9
 
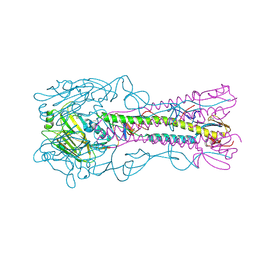 | | The structure of hemagglutinin from a H6N1 influenza virus (A/Taiwan/2/2013) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HA1, HA2, ... | | Authors: | Wang, F, Qi, J, Bi, Y, Zhang, W, Wang, M, Wang, M, Liu, J, Yan, J, Shi, Y, Gao, G.F. | | Deposit date: | 2015-03-23 | | Release date: | 2016-04-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structure of hemagglutinin from a H6N1 influenza virus (A/Taiwan/2/2013) at 2.6 Angstroms resolution
To Be Published
|
|
9BQ0
 
 | |
4YY7
 
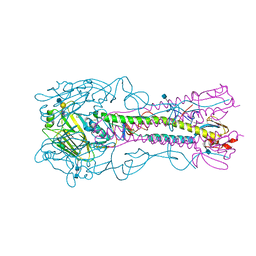 | | The structure of hemagglutinin from a H6N1 influenza virus (A/chicken/Taiwan/A2837/2013) in complex with avian receptor analog 3'SLNLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HA1, ... | | Authors: | Wang, F, Qi, J, Bi, Y, Zhang, W, Wang, M, Wang, M, Liu, J, Yan, J, Shi, Y, Gao, G.F. | | Deposit date: | 2015-03-23 | | Release date: | 2016-03-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structure of hemagglutinin from a H6N1 influenza virus (A/chicken/Taiwan/A2837/2013)
To Be Published
|
|
4YY0
 
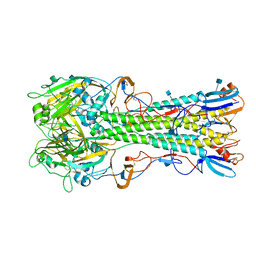 | | The structure of hemagglutinin from a H6N1 influenza virus (A/chicken/Taiwan/A2837/2013) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HA1, HA2 | | Authors: | Wang, F, Qi, J, Bi, Y, Zhang, W, Wang, M, Wang, M, Liu, J, Yan, J, Shi, Y, Gao, G.F. | | Deposit date: | 2015-03-23 | | Release date: | 2016-03-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structure of hemagglutinin from a H6N1 influenza virus (A/chicken/Taiwan/A2837/2013)
To Be Published
|
|
2LSP
 
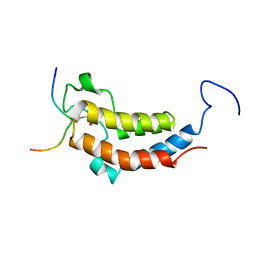 | | solution structures of BRD4 second bromodomain with NF-kB-K310ac peptide | | Descriptor: | Bromodomain-containing protein 4, NF-kB-K310ac peptide | | Authors: | Zhang, G, Liu, R, Zhong, Y, Plotnikov, A.N, Zhang, W, Rusinova, E, Gerona-Nevarro, G, Moshkina, N, Joshua, J, Chuang, P.Y, Ohlmeyer, M, He, J, Zhou, M.-M. | | Deposit date: | 2012-05-03 | | Release date: | 2012-07-18 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Down-regulation of NF-kappa B transcriptional activity in HIV-associated kidney disease by BRD4 inhibition.
J.Biol.Chem., 287, 2012
|
|
4NS5
 
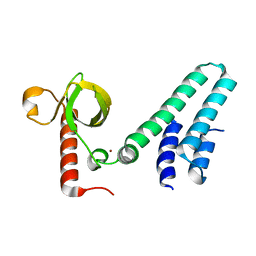 | | Crystal structure of human BS69 Bromo-Zinc finger-PWWP | | Descriptor: | ZINC ION, Zinc finger MYND domain-containing protein 11 | | Authors: | Wang, J.C, Qin, S, Li, F.D, Li, S, Zhang, W, Wu, J.H, Shi, Y.Y. | | Deposit date: | 2013-11-28 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human BS69 Bromo-ZnF-PWWP reveals its role in H3K36me3 nucleosome binding.
Cell Res., 24, 2014
|
|
