1RNI
 
 | | Bifunctional DNA primase/polymerase domain of ORF904 from the archaeal plasmid pRN1 | | Descriptor: | ORF904, ZINC ION | | Authors: | Lipps, G, Weinzierl, A.O, von Scheven, G, Buchen, C, Cramer, P. | | Deposit date: | 2003-12-01 | | Release date: | 2004-01-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of a bifunctional DNA primase-polymerase
Nat.Struct.Mol.Biol., 11, 2004
|
|
1RO2
 
 | | Bifunctional DNA primase/polymerase domain of ORF904 from the archaeal plasmid pRN1- Triple mutant F50M/L107M/L110M manganese soak | | Descriptor: | MANGANESE (II) ION, ZINC ION, hypothetical protein ORF904 | | Authors: | Lipps, G, Weinzierl, A.O, von Scheven, G, Buchen, C, Cramer, P. | | Deposit date: | 2003-12-01 | | Release date: | 2004-01-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of a bifunctional DNA primase-polymerase
Nat.Struct.Mol.Biol., 11, 2004
|
|
8GWR
 
 | | Near full length Kidney type Glutaminase in complex with 2,2-Dimethyl-2,3-Dihydrobenzo[a] Phenanthridin-4(1H)-one (DDP) | | Descriptor: | 2,2-dimethyl-1,3-dihydrobenzo[a]phenanthridin-4-one, Glutaminase kidney isoform, mitochondrial | | Authors: | Shankar, S, Jobichen, C, Sivaraman, J. | | Deposit date: | 2022-09-17 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | A novel allosteric site employs a conserved inhibition mechanism in human kidney-type glutaminase.
Febs J., 290, 2023
|
|
8YUE
 
 | | Crystal structure of the kinesin-14 motor protein from Drosophila melanogaster | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PHOSPHATE ION, Protein claret segregational | | Authors: | Wei, Y, Jobichen, C, Imasaki, T, Nitta, R, Wang, M.Y, Sivaraman, J, Endow, S.A. | | Deposit date: | 2024-03-27 | | Release date: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal structure of a kinesin-related protein from Drosophila melanogaster
To Be Published
|
|
8HBC
 
 | | Crystal structure of the CysR-CTLD3 fragment of human DEC205 | | Descriptor: | Lymphocyte antigen 75 | | Authors: | Kong, D, Yu, B, Hu, Z, Cheng, C, Cao, L, He, Y. | | Deposit date: | 2022-10-28 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Interaction of human dendritic cell receptor DEC205/CD205 with keratins.
J.Biol.Chem., 300, 2024
|
|
6JLI
 
 | | Crystal structure of CTLD7 domain of human PLA2R | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Secretory phospholipase A2 receptor | | Authors: | Yu, B, Hu, Z, Kong, D, Cheng, C, He, Y. | | Deposit date: | 2019-03-06 | | Release date: | 2019-07-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.778 Å) | | Cite: | Crystal structure of the CTLD7 domain of human M-type phospholipase A2 receptor.
J.Struct.Biol., 207, 2019
|
|
7E0W
 
 | | Crystal Structure of BCH domain from S. pombe | | Descriptor: | Putative Rho GTPase-activating protein C1565.02c, TETRAETHYLENE GLYCOL | | Authors: | Chichili, V.P.R, Jobichen, C, Sivaraman, J. | | Deposit date: | 2021-01-28 | | Release date: | 2021-03-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A novel intertwined anti-parallel dimeric structure of scaffold BCH domain regulates RhoA and RhoGAP functions
Proc.Natl.Acad.Sci.USA, 2021
|
|
7E4U
 
 | |
7E4V
 
 | |
7E50
 
 | | Crystal structure of human microplasmin in complex with kazal-type inhibitor AaTI | | Descriptor: | AAEL006007-PA, GLYCEROL, Plasminogen, ... | | Authors: | Varsha, A.W, Jobichen, C, Mok, Y.K. | | Deposit date: | 2021-02-16 | | Release date: | 2022-02-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of Aedes aegypti trypsin inhibitor in complex with mu-plasmin reveals role for scaffold stability in Kazal-type serine protease inhibitor.
Protein Sci., 31, 2022
|
|
7EQZ
 
 | | Crystal structure of Carboxypeptidase B complexed with Potato Carboxypeptidase Inhibitor | | Descriptor: | Carboxypeptidase B, GLYCINE, Metallocarboxypeptidase inhibitor, ... | | Authors: | Choong, Y.K, Gavor, E, Jobichen, C, Sivaraman, J. | | Deposit date: | 2021-05-05 | | Release date: | 2021-11-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Aedes aegypti carboxypeptidase B1-inhibitor complex uncover the disparity between mosquito and non-mosquito insect carboxypeptidase inhibition mechanism.
Protein Sci., 30, 2021
|
|
2O4X
 
 | | Crystal structure of human P100 tudor domain | | Descriptor: | Staphylococcal nuclease domain-containing protein 1 | | Authors: | Shaw, N, Zhao, M, Cheng, C, Xu, H, Yang, J, Silvennoinen, O, Rao, Z, Wang, B.C, Liu, Z.J. | | Deposit date: | 2006-12-05 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human P100 tudor domain
To be Published
|
|
3KET
 
 | | Crystal structure of a Rex-family transcriptional regulatory protein from Streptococcus agalactiae bound to a palindromic operator | | Descriptor: | DNA (5'-D(*AP*AP*TP*TP*GP*TP*GP*AP*AP*AP*T)-3'), DNA (5'-D(P*AP*TP*TP*TP*CP*AP*CP*AP*AP*TP*T)-3'), MAGNESIUM ION, ... | | Authors: | Thiyagarajan, S, Logan, D, von Wachenfeldt, C. | | Deposit date: | 2009-10-26 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | NAD+ pool depletion as a signal for the Rex regulon involved in Streptococcus agalactiae virulence.
Plos Pathog., 17, 2021
|
|
3KEO
 
 | | Crystal Structure of a Rex-family transcriptional regulatory protein from Streptococcus agalactiae complexed with NAD+ | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Thiyagarajan, S, Logan, D, von Wachenfeldt, C. | | Deposit date: | 2009-10-26 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | NAD+ pool depletion as a signal for the Rex regulon involved in Streptococcus agalactiae virulence.
Plos Pathog., 17, 2021
|
|
3HY6
 
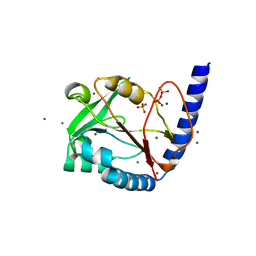 | | Structure of human MTHFS with ADP | | Descriptor: | 5-formyltetrahydrofolate cyclo-ligase, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wu, D, Li, Y, Song, G, Cheng, C, Shaw, N, Liu, Z.-J. | | Deposit date: | 2009-06-22 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the inhibition of human 5,10-methenyltetrahydrofolate synthetase by N10-substituted folate analogues
Cancer Res., 69, 2009
|
|
3HXT
 
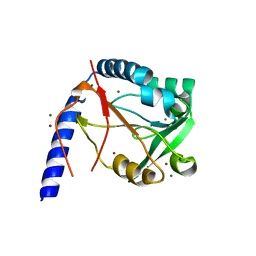 | | Structure of human MTHFS | | Descriptor: | 5-formyltetrahydrofolate cyclo-ligase, MAGNESIUM ION, NICKEL (II) ION | | Authors: | Wu, D, Li, Y, Song, G, Cheng, C, Zhang, R, Joachimiak, A, Shaw, N, Liu, Z.-J. | | Deposit date: | 2009-06-22 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the inhibition of human 5,10-methenyltetrahydrofolate synthetase by N10-substituted folate analogues
Cancer Res., 69, 2009
|
|
3HY3
 
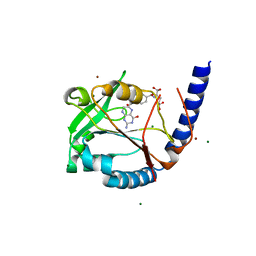 | | Structure of human MTHFS with 10-formyltetrahydrofolate | | Descriptor: | 5-formyltetrahydrofolate cyclo-ligase, MAGNESIUM ION, N-({4-[{[(2R,4S,4aR,6S,8aS)-2-amino-4-hydroxydecahydropteridin-6-yl]methyl}(formyl)amino]phenyl}carbonyl)-D-glutamic acid, ... | | Authors: | Wu, D, Li, Y, Song, G, Cheng, C, Shaw, N, Liu, Z.-J. | | Deposit date: | 2009-06-22 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the inhibition of human 5,10-methenyltetrahydrofolate synthetase by N10-substituted folate analogues
Cancer Res., 69, 2009
|
|
3HY4
 
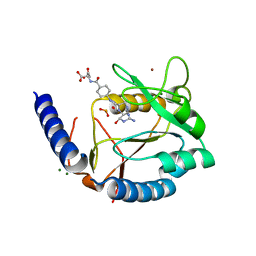 | | Structure of human MTHFS with N5-iminium phosphate | | Descriptor: | 5-formyltetrahydrofolate cyclo-ligase, MAGNESIUM ION, N-({trans-4-[({(2R,4R,4aS,6S,8aS)-2-amino-4-hydroxy-5-[(phosphonooxy)methyl]decahydropteridin-6-yl}methyl)amino]cyclohexyl}carbonyl)-L-glutamic acid, ... | | Authors: | Wu, D, Li, Y, Song, G, Cheng, C, Shaw, N, Liu, Z.-J. | | Deposit date: | 2009-06-22 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.795 Å) | | Cite: | Structural basis for the inhibition of human 5,10-methenyltetrahydrofolate synthetase by N10-substituted folate analogues
Cancer Res., 69, 2009
|
|
3KEQ
 
 | |
2HQE
 
 | | Crystal structure of human P100 Tudor domain: Large fragment | | Descriptor: | P100 Co-activator tudor domain | | Authors: | Shah, N, Zhao, M, Cheng, C, Xu, H, Yang, J, Silvennoinen, O, Liu, Z.J, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-07-18 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a large fragment of the Human P100 Tudor Domain
To be Published
|
|
8DI5
 
 | | Cryo-EM structure of SARS-CoV-2 Beta (B.1.351) spike protein in complex with VH domain F6 (focused refinement of RBD and VH F6) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, VH F6 | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Subramaniam, S. | | Deposit date: | 2022-06-28 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Potent and broad neutralization of SARS-CoV-2 variants of concern (VOCs) including omicron sub-lineages BA.1 and BA.2 by biparatopic human VH domains.
Iscience, 25, 2022
|
|
4MPY
 
 | | 1.85 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) from Staphylococcus aureus (IDP00699) in complex with NAD+ | | Descriptor: | Betaine aldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION | | Authors: | Halavaty, A.S, Minasov, G, Shuvalova, L, Winsor, J, Peterson, S.N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-14 | | Release date: | 2013-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-based mutational studies of substrate inhibition of betaine aldehyde dehydrogenase BetB from Staphylococcus aureus.
Appl.Environ.Microbiol., 80, 2014
|
|
4MPB
 
 | | 1.7 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) from Staphylococcus aureus | | Descriptor: | Betaine aldehyde dehydrogenase, CHLORIDE ION, MAGNESIUM ION | | Authors: | Halavaty, A.S, Shuvalova, L, Minasov, G, Dubrovska, I, Winsor, J, Peterson, S.N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-12 | | Release date: | 2013-09-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based mutational studies of substrate inhibition of betaine aldehyde dehydrogenase BetB from Staphylococcus aureus.
Appl.Environ.Microbiol., 80, 2014
|
|
3FDR
 
 | | Crystal structure of TDRD2 | | Descriptor: | Tudor and KH domain-containing protein | | Authors: | Amaya, M.F, Adams, M.A, Guo, Y, Li, Y, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-26 | | Release date: | 2009-01-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mouse Piwi interactome identifies binding mechanism of Tdrkh Tudor domain to arginine methylated Miwi
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2MC1
 
 | | Solution structure of the Vav1 SH2 domain complexed with a Syk-derived singly phosphorylated peptide | | Descriptor: | Proto-oncogene vav, Tyrosine-protein kinase SYK | | Authors: | Chen, C, Piraner, D, Gorenstein, N.M, Geahlen, R.L, Post, C.B. | | Deposit date: | 2013-08-13 | | Release date: | 2013-08-28 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Differential recognition of syk-binding sites by each of the two phosphotyrosine-binding pockets of the Vav SH2 domain.
Biopolymers, 99, 2013
|
|
