6UUM
 
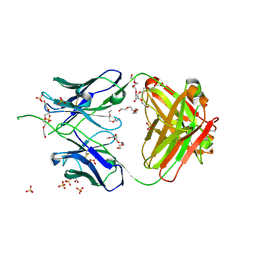 | | Crystal structure of antibody 438-B11 DSS mutant (Cys98A-Cys100aA) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, B11 DSS Fab Heavy Chain, ... | | Authors: | Kumar, S, Wilson, I.A. | | Deposit date: | 2019-10-30 | | Release date: | 2020-09-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A V H 1-69 antibody lineage from an infected Chinese donor potently neutralizes HIV-1 by targeting the V3 glycan supersite.
Sci Adv, 6, 2020
|
|
6UUL
 
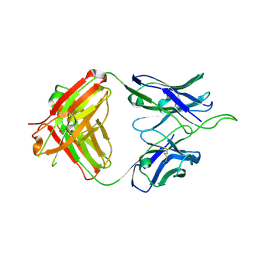 | | Crystal structure of broad and potent HIV-1 neutralizing antibody 438-D5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, D5 Fab Heavy Chain, ... | | Authors: | Kumar, S, Wilson, I.A. | | Deposit date: | 2019-10-30 | | Release date: | 2020-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | A V H 1-69 antibody lineage from an infected Chinese donor potently neutralizes HIV-1 by targeting the V3 glycan supersite.
Sci Adv, 6, 2020
|
|
6UYC
 
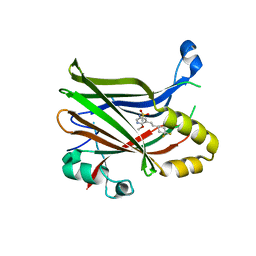 | | Crystal structure of TEAD2 bound to Compound 2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-{5-[(E)-2-(4,4-difluorocyclohexyl)ethenyl]-6-methoxypyridin-3-yl}methanesulfonamide, Transcriptional enhancer factor TEF-4 | | Authors: | Noland, C.L, Holden, J.K, Crawford, J.J, Zbieg, J.R, Cunningham, C.N. | | Deposit date: | 2019-11-12 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.658 Å) | | Cite: | Small Molecule Dysregulation of TEAD Lipidation Induces a Dominant-Negative Inhibition of Hippo Pathway Signaling.
Cell Rep, 31, 2020
|
|
6UUH
 
 | |
6UYB
 
 | | Crystal structure of TEAD2 bound to Compound 1 | | Descriptor: | (3R,4R)-1-{3-[(E)-2-(4-chlorophenyl)ethenyl]-4-methoxy-5-methylphenyl}-3,4-dihydroxypyrrolidin-2-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ... | | Authors: | Noland, C.L, Holden, J.K, Crawford, J.J, Zbieg, J.R, Cunningham, C.N. | | Deposit date: | 2019-11-12 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.543 Å) | | Cite: | Small Molecule Dysregulation of TEAD Lipidation Induces a Dominant-Negative Inhibition of Hippo Pathway Signaling.
Cell Rep, 31, 2020
|
|
7Y3A
 
 | | Crystal structure of TRIM7 bound to 2C | | Descriptor: | E3 ubiquitin-protein ligase TRIM7,E3 ubiquitin-protein ligase TRIM7,E3 ubiquitin-protein ligase TRIM7,TRIM7-2C | | Authors: | Dong, C, Yan, X. | | Deposit date: | 2022-06-10 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | C-terminal glutamine acts as a C-degron targeted by E3 ubiquitin ligase TRIM7.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Y3B
 
 | | Crystal structure of TRIM7 bound to GN1 | | Descriptor: | E3 ubiquitin-protein ligase TRIM7,TRIM7-GN1 | | Authors: | Dong, C, Yan, X. | | Deposit date: | 2022-06-10 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | C-terminal glutamine acts as a C-degron targeted by E3 ubiquitin ligase TRIM7.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Y3C
 
 | | Crystal structure of TRIM7 bound to RACO-1 | | Descriptor: | E3 ubiquitin-protein ligase TRIM7,E3 ubiquitin-protein ligase TRIM7,TRIM7-RACO-1 | | Authors: | Dong, C, Yan, X. | | Deposit date: | 2022-06-10 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | C-terminal glutamine acts as a C-degron targeted by E3 ubiquitin ligase TRIM7.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6V6W
 
 | |
4Z69
 
 | | Human serum albumin complexed with palmitic acid and diclofenac | | Descriptor: | 2-[2,6-DICHLOROPHENYL)AMINO]BENZENEACETIC ACID, PALMITIC ACID, PENTADECANOIC ACID, ... | | Authors: | Zhang, Y, Yang, F. | | Deposit date: | 2015-04-04 | | Release date: | 2016-01-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | Structural basis of non-steroidal anti-inflammatory drug diclofenac binding to human serum albumin.
Chem.Biol.Drug Des., 86, 2015
|
|
8ZU2
 
 | | Crystal Structure of Human Myt1 Kinase domain Bounded with compound 8g | | Descriptor: | 2-azanyl-5-[2-(1,4-diazepan-1-yl)pyridin-4-yl]-3-(2,6-dimethyl-3-oxidanyl-phenyl)benzamide, GLYCINE, Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase | | Authors: | Zhang, Z.M, Zhou, Z.Q. | | Deposit date: | 2024-06-07 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.79888582 Å) | | Cite: | Structure-Based Drug Design of 2-Amino-[1,1'-biphenyl]-3-carboxamide Derivatives as Selective PKMYT1 Inhibitors for the Treatment of CCNE1 -Amplified Breast Cancer.
J.Med.Chem., 67, 2024
|
|
8ZUL
 
 | | Crystal Structure of Human Myt1 Kinase domain Bounded with compound 8m | | Descriptor: | 2-azanyl-5-[2-[(3~{R})-3-azanylpyrrolidin-1-yl]pyridin-4-yl]-3-(2,6-dimethyl-3-oxidanyl-phenyl)benzamide, Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase | | Authors: | Zhang, Z.M, Zhou, Z.Q. | | Deposit date: | 2024-06-09 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.80026162 Å) | | Cite: | Structure-Based Drug Design of 2-Amino-[1,1'-biphenyl]-3-carboxamide Derivatives as Selective PKMYT1 Inhibitors for the Treatment of CCNE1 -Amplified Breast Cancer.
J.Med.Chem., 67, 2024
|
|
8ZTX
 
 | | Crystal Structure of Human Myt1 Kinase domain Bounded with compound 6b | | Descriptor: | 2-azanyl-3-(2,6-dimethyl-3-oxidanyl-phenyl)-5-pyridin-4-yl-benzamide, Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase | | Authors: | Zhang, Z.M, Zhou, Z.Q. | | Deposit date: | 2024-06-07 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.70033228 Å) | | Cite: | Structure-Based Drug Design of 2-Amino-[1,1'-biphenyl]-3-carboxamide Derivatives as Selective PKMYT1 Inhibitors for the Treatment of CCNE1 -Amplified Breast Cancer.
J.Med.Chem., 67, 2024
|
|
8ZUD
 
 | | Crystal Structure of Human Myt1 Kinase domain Bounded with compound 8f | | Descriptor: | 2-azanyl-3-(2,6-dimethyl-3-oxidanyl-phenyl)-5-(2-morpholin-4-ylpyridin-4-yl)benzamide, Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase | | Authors: | Zhang, Z.M, Zhou, Z.Q. | | Deposit date: | 2024-06-08 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.50510085 Å) | | Cite: | Structure-Based Drug Design of 2-Amino-[1,1'-biphenyl]-3-carboxamide Derivatives as Selective PKMYT1 Inhibitors for the Treatment of CCNE1 -Amplified Breast Cancer.
J.Med.Chem., 67, 2024
|
|
7DOI
 
 | |
7DOK
 
 | |
7CR5
 
 | |
8VHG
 
 | | Structure of the BMAL1/HIF2A heterodimer in Complex with DNA | | Descriptor: | Basic helix-loop-helix ARNT-like protein 1, Endothelial PAS domain-containing protein 1, Forward strand DNA containing HRE motif, ... | | Authors: | Li, T, Tsai, K.L. | | Deposit date: | 2024-01-01 | | Release date: | 2025-02-12 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | BMAL1-HIF2A heterodimer modulates circadian variations of myocardial injury.
Nature, 641, 2025
|
|
7DE1
 
 | |
8XAJ
 
 | | Cryo-EM structure of OSCA1.2-liposome-inside-in open state | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Calcium permeable stress-gated cation channel 1 | | Authors: | Zhang, Y, Han, Y. | | Deposit date: | 2023-12-04 | | Release date: | 2024-04-10 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Mechanical activation opens a lipid-lined pore in OSCA ion channels.
Nature, 628, 2024
|
|
8ZOV
 
 | |
8ZO3
 
 | |
8ZPO
 
 | |
8K9D
 
 | | Structure of human Caprin-2 HR1 domain | | Descriptor: | Caprin-2 | | Authors: | Song, X.M. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-21 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural insights into the Caprin-2 HR1 domain in canonical Wnt signaling.
J.Biol.Chem., 300, 2024
|
|
5Z78
 
 | | Structure of TIRR/53BP1 complex | | Descriptor: | TP53-binding protein 1, Tudor-interacting repair regulator protein | | Authors: | Dai, Y.X, Shan, S. | | Deposit date: | 2018-01-27 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.762 Å) | | Cite: | Structural basis for recognition of 53BP1 tandem Tudor domain by TIRR
Nat Commun, 9, 2018
|
|
