7WKD
 
 | | TRH-TRHR G protein complex | | Descriptor: | Gq, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, Y, Cai, H, You, C, He, X, Yuan, Q, Jiang, H, Cheng, X, Jiang, Y, Xu, H.E. | | Deposit date: | 2022-01-09 | | Release date: | 2022-06-15 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural insights into ligand binding and activation of the human thyrotropin-releasing hormone receptor.
Cell Res., 32, 2022
|
|
7XO7
 
 | | SARS-CoV-2 Omicron BA.2 Variant Spike Trimer with two human ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, CHLORIDE ION, ... | | Authors: | Xu, Y, Wu, C, Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2022-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins.
Cell Res., 32, 2022
|
|
7XO8
 
 | | SARS-CoV-2 Omicron BA.2 Variant Spike Trimer with three human ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, CHLORIDE ION, ... | | Authors: | Xu, Y, Wu, C, Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2022-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins.
Cell Res., 32, 2022
|
|
7XO4
 
 | | SARS-CoV-2 Omicron BA.1 Variant Spike Trimer with two mouse ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Xu, Y, Wu, C, Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2022-04-30 | | Release date: | 2022-06-15 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins.
Cell Res., 32, 2022
|
|
7XO6
 
 | | SARS-CoV-2 Omicron BA.1 Variant RBD with mouse ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Xu, Y, Wu, C, Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2022-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins.
Cell Res., 32, 2022
|
|
7XOA
 
 | | SARS-CoV-2 Omicron BA.2 Variant Spike Trimer with one mouse ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Xu, Y, Wu, C, Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2022-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins.
Cell Res., 32, 2022
|
|
7XOC
 
 | | SARS-CoV-2 Omicron BA.2 Variant RBD complexed with mouse ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Xu, Y, Wu, C, Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2022-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins.
Cell Res., 32, 2022
|
|
7XOB
 
 | | SARS-CoV-2 Omicron BA.2 Variant Spike Trimer with two mouse ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Xu, Y, Wu, C, Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2022-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins.
Cell Res., 32, 2022
|
|
7XO5
 
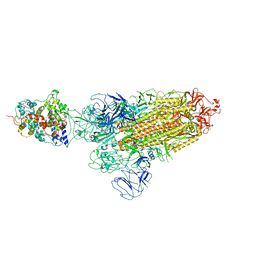 | | SARS-CoV-2 Omicron BA.1 Variant Spike Trimer with one mouse ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Xu, Y, Wu, C, Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2022-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins.
Cell Res., 32, 2022
|
|
7XOD
 
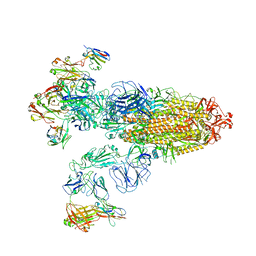 | | SARS-CoV-2 Omicron BA.2 Variant Spike Trimer with three JMB2002 Fab Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of JMB2002 Fab, ... | | Authors: | Xu, Y, Wu, C, Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2022-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins.
Cell Res., 32, 2022
|
|
7XO9
 
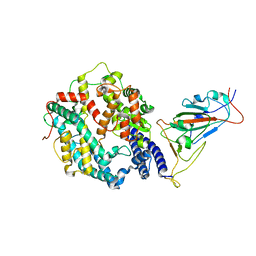 | | SARS-CoV-2 Omicron BA.2 Variant RBD complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, CHLORIDE ION, ... | | Authors: | Xu, Y, Wu, C, Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2022-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins.
Cell Res., 32, 2022
|
|
7XZ5
 
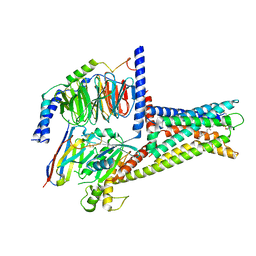 | | GPR119-Gs-LPC complex | | Descriptor: | (4R,7R,18E)-4,7-dihydroxy-N,N,N-trimethyl-10-oxo-3,5,9-trioxa-4-phosphaheptacos-18-en-1-aminium 4-oxide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, P, Huang, S, Jiang, Y, Xu, H.E. | | Deposit date: | 2022-06-02 | | Release date: | 2022-08-24 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural identification of lysophosphatidylcholines as activating ligands for orphan receptor GPR119.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7XZ6
 
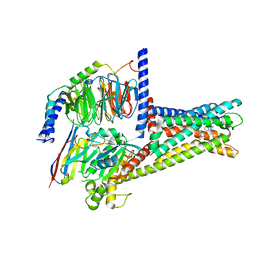 | | GPR119-Gs-APD668 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Xu, P, Huang, S, Jiang, Y, Xu, H.E. | | Deposit date: | 2022-06-02 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural identification of lysophosphatidylcholines as activating ligands for orphan receptor GPR119.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7BV2
 
 | | The nsp12-nsp7-nsp8 complex bound to the template-primer RNA and triphosphate form of Remdesivir(RTP) | | Descriptor: | MAGNESIUM ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Yin, W, Mao, C, Luan, X, Shen, D, Shen, Q, Su, H, Wang, X, Zhou, F, Zhao, W, Gao, M, Chang, S, Xie, Y.C, Tian, G, Jiang, H.W, Tao, S.C, Shen, J, Jiang, Y, Jiang, H, Xu, Y, Zhang, S, Zhang, Y, Xu, H.E. | | Deposit date: | 2020-04-09 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir.
Science, 368, 2020
|
|
7BV1
 
 | | Cryo-EM structure of the apo nsp12-nsp7-nsp8 complex | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA-directed RNA polymerase, ... | | Authors: | Yin, W, Mao, C, Luan, X, Shen, D, Shen, Q, Su, H, Wang, X, Zhou, F, Zhao, W, Gao, M, Chang, S, Xie, Y.C, Tian, G, Jiang, H.W, Tao, S.C, Shen, J, Jiang, Y, Jiang, H, Xu, Y, Zhang, S, Zhang, Y, Xu, H.E. | | Deposit date: | 2020-04-09 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir.
Science, 368, 2020
|
|
4PXQ
 
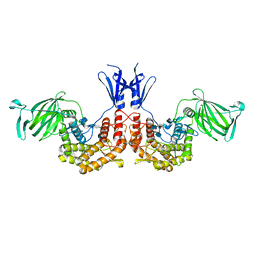 | | Crystal structure of D-glucuronyl C5-epimerase in complex with heparin hexasaccharide | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, D-glucuronyl C5 epimerase B | | Authors: | Ke, J, Qin, Y, Gu, X, Tan, J, Brunzelle, J.S, Xu, H.E, Ding, K. | | Deposit date: | 2014-03-24 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Functional Study of d-Glucuronyl C5-epimerase.
J.Biol.Chem., 290, 2015
|
|
7WBJ
 
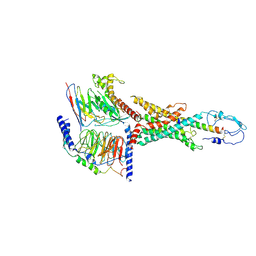 | | Cryo-EM structure of N-terminal modified human vasoactive intestinal polypeptide receptor 2 (VIP2R) in complex with PACAP27 and Gs | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Xu, Y.N, Feng, W.B, Zhou, Q.T, Liang, A.Y, Li, J, Dai, A.T, Zhao, F.H, Yan, J.H, Chen, C.W, Li, H, Zhao, L.H, Xia, T, Jiang, Y, Xu, H.E, Yang, D.H, Wang, M.W. | | Deposit date: | 2021-12-16 | | Release date: | 2022-05-18 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | A distinctive ligand recognition mechanism by the human vasoactive intestinal polypeptide receptor 2.
Nat Commun, 13, 2022
|
|
7VQX
 
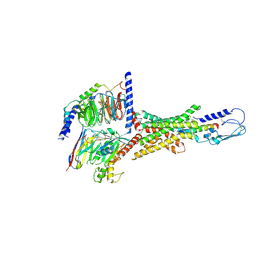 | | Cryo-EM structure of human vasoactive intestinal polypeptide receptor 2 (VIP2R) in complex with PACAP27 and Gs | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Xu, Y.N, Feng, W.B, Zhou, Q.T, Liang, A.Y, Li, J, Dai, A.T, Zhao, F.H, Yan, J.H, Chen, C.W, Li, H, Zhao, L.H, Xia, T, Jiang, Y, Xu, H.E, Yang, D.H, Wang, M.W. | | Deposit date: | 2021-10-21 | | Release date: | 2022-05-18 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | A distinctive ligand recognition mechanism by the human vasoactive intestinal polypeptide receptor 2.
Nat Commun, 13, 2022
|
|
4P6X
 
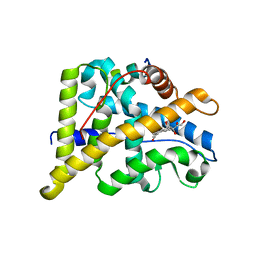 | | Crystal Structure of cortisol-bound glucocorticoid receptor ligand binding domain | | Descriptor: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, Glucocorticoid receptor, Nuclear receptor coactivator 2 | | Authors: | He, Y, Zhou, X.E, Tolbert, W.D, Powell, K, Melcher, K, Xu, H.E. | | Deposit date: | 2014-03-25 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures and mechanism for the design of highly potent glucocorticoids.
Cell Res., 24, 2014
|
|
7VBH
 
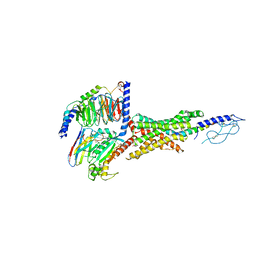 | | Cryo-EM structure of the GIPR/GLP-1R/GCGR triagonist peptide 20-bound human GLP-1R-Gs complex | | Descriptor: | Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, F.H, Zhou, Q.T, Cong, Z.T, Hang, K.N, Zou, X.Y, Zhang, C, Chen, Y, Dai, A.T, Liang, A.Y, Ming, Q.Q, Wang, M, Chen, L.N, Xu, P.Y, Chang, R.L, Feng, W.B, Xia, T, Zhang, Y, Wu, B.L, Yang, D.H, Zhao, L.H, Xu, H.E, Wang, M.W. | | Deposit date: | 2021-08-31 | | Release date: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into multiplexed pharmacological actions of tirzepatide and peptide 20 at the GIP, GLP-1 or glucagon receptors.
Nat Commun, 13, 2022
|
|
7V35
 
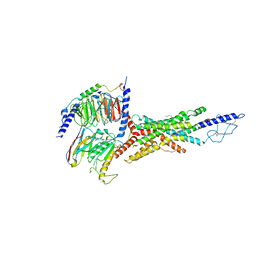 | | Cryo-EM structure of the GIPR/GLP-1R/GCGR triagonist peptide 20-bound human GCGR-Gs complex | | Descriptor: | Glucagon receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, F.h, Zhou, Q.T, Cong, Z.T, Hang, K.N, Zou, X.Y, Zhang, C, Chen, Y, Dai, A.T, Liang, A.Y, Ming, Q.Q, Wang, M, Chen, L.N, Xu, P.Y, Chang, R.L, Feng, W.B, Xia, T, Zhang, Y, Wu, B.L, Yang, D.H, Zhao, L.H, Xu, H.E, Wang, M.W. | | Deposit date: | 2021-08-10 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into multiplexed pharmacological actions of tirzepatide and peptide 20 at the GIP, GLP-1 or glucagon receptors.
Nat Commun, 13, 2022
|
|
7VAB
 
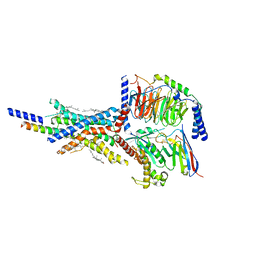 | | Cryo-EM structure of the non-acylated tirzepatide (LY3298176)-bound human GIPR-Gs complex | | Descriptor: | CHOLESTEROL, Gastric inhibitory polypeptide receptor,Gastric inhibitory polypeptide receptor,Gastric inhibitory polypeptide receptor,human glucose-dependent insulinotropic polypeptide receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhao, F.H, Zhou, Q.T, Cong, Z.T, Hang, K.N, Zou, X.Y, Zhang, C, Chen, Y, Dai, A.T, Liang, A.Y, Ming, Q.Q, Wang, M, Chen, L.N, Xu, P.Y, Chang, R.L, Feng, W.B, Xia, T, Zhang, Y, Wu, B.L, Yang, D.H, Zhao, L.H, Xu, H.E, Wang, M.W. | | Deposit date: | 2021-08-28 | | Release date: | 2022-03-02 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into multiplexed pharmacological actions of tirzepatide and peptide 20 at the GIP, GLP-1 or glucagon receptors.
Nat Commun, 13, 2022
|
|
7VBI
 
 | | Cryo-EM structure of the non-acylated tirzepatide (LY3298176)-bound human GLP-1R-Gs complex | | Descriptor: | Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, F.H, Zhou, Q.T, Cong, Z.T, Hang, K.N, Zou, X.Y, Zhang, C, Chen, Y, Dai, A.T, Liang, A.Y, Ming, Q.Q, Wang, M, Chen, L.N, Xu, P.Y, Chang, R.L, Feng, W.B, Xia, T, Zhang, Y, Wu, B.L, Yang, D.H, Zhao, L.H, Xu, H.E, Wang, M.W. | | Deposit date: | 2021-08-31 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into multiplexed pharmacological actions of tirzepatide and peptide 20 at the GIP, GLP-1 or glucagon receptors.
Nat Commun, 13, 2022
|
|
7E2Y
 
 | | Serotonin-bound Serotonin 1A (5-HT1A) receptor-Gi protein complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, P, Huang, S, Zhang, H, Mao, C, Zhou, X.E, Shen, D.D, Jiang, Y, Zhang, Y, Xu, H.E. | | Deposit date: | 2021-02-07 | | Release date: | 2021-04-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the lipid and ligand regulation of serotonin receptors.
Nature, 592, 2021
|
|
7E33
 
 | | Serotonin 1E (5-HT1E) receptor-Gi protein complex | | Descriptor: | 3-(1-methylpiperidin-4-yl)-1H-indol-5-ol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, P, Huang, S, Zhang, H, Mao, C, Zhou, X.E, Shen, D.D, Jiang, Y, Zhang, Y, Xu, H.E. | | Deposit date: | 2021-02-07 | | Release date: | 2021-04-14 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into the lipid and ligand regulation of serotonin receptors.
Nature, 592, 2021
|
|
