6C6J
 
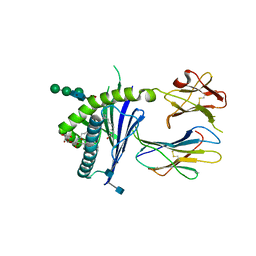 | | Structure of glycolipid aGSA[8,P5p] in complex with mouse CD1d | | Descriptor: | (5R,6S,7S)-5,6-dihydroxy-7-(octanoylamino)-N-(4-pentylphenyl)-8-{[(2S,3R,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)te trahydro-2H-pyran-2-yl]oxy}octanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Zajonc, D.M, Wang, J. | | Deposit date: | 2018-01-18 | | Release date: | 2019-01-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A molecular switch in mouse CD1d modulates natural killer T cell activation by alpha-galactosylsphingamides.
J.Biol.Chem., 294, 2019
|
|
6M1H
 
 | | CryoEM structure of human PAC1 receptor in complex with maxadilan | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Song, X, Wang, J, Zhang, D, Wang, H.W, Ma, Y. | | Deposit date: | 2020-02-26 | | Release date: | 2020-03-11 | | Last modified: | 2020-05-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of PAC1 receptor reveal ligand binding mechanism.
Cell Res., 30, 2020
|
|
6C69
 
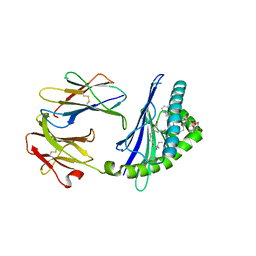 | | Structure of glycolipid aGSA[12,6P] in complex with mouse CD1d | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Zajonc, D.M, Wang, J. | | Deposit date: | 2018-01-18 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.937 Å) | | Cite: | A molecular switch in mouse CD1d modulates natural killer T cell activation by alpha-galactosylsphingamides.
J.Biol.Chem., 294, 2019
|
|
2FG5
 
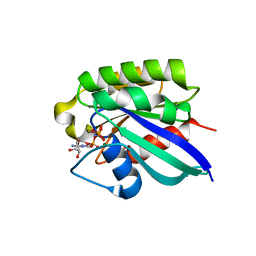 | | Crystal structure of human RAB31 in complex with a GTP analogue | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein Rab-31 | | Authors: | Tempel, W, Wang, J, Ismail, S, Arrowsmith, C, Edwards, A, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-12-21 | | Release date: | 2006-01-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Crystal structure of human RAB31 in complex with a GTP analogue
To be Published
|
|
7LHL
 
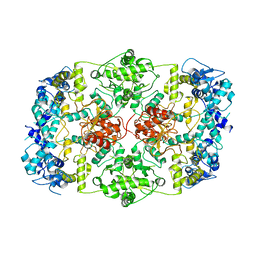 | |
7LU6
 
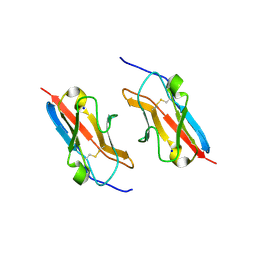 | | Crystal structure of the mouse Kirrel3 D1 homodimer | | Descriptor: | Kin of IRRE-like protein 3, SODIUM ION | | Authors: | Roman, C.A, Pak, J.S, Wang, J, Ozkan, E. | | Deposit date: | 2021-02-21 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular and structural basis of olfactory sensory neuron axon coalescence by Kirrel receptors.
Cell Rep, 37, 2021
|
|
7LTW
 
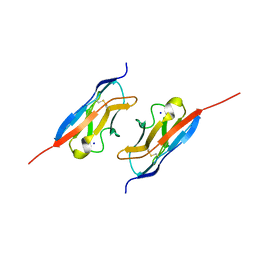 | | Crystal structure of the mouse Kirrel2 D1 homodimer | | Descriptor: | Kin of IRRE-like protein 2, SODIUM ION | | Authors: | Roman, C.A, Pak, J.S, Wang, J, Ozkan, E. | | Deposit date: | 2021-02-20 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular and structural basis of olfactory sensory neuron axon coalescence by Kirrel receptors.
Cell Rep, 37, 2021
|
|
8IFF
 
 | | Cryo-EM structure of Arabidopsis phytochrome A. | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome A | | Authors: | Ma, L, Zhou, C, Wang, J, Guan, Z, Yin, P. | | Deposit date: | 2023-02-17 | | Release date: | 2023-08-02 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Plant phytochrome A in the Pr state assembles as an asymmetric dimer.
Cell Res., 33, 2023
|
|
7UW6
 
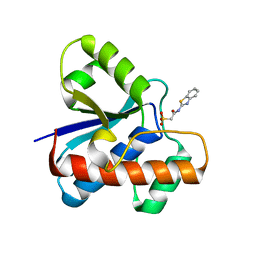 | | The co-crystal structure of low molecular weight protein tyrosine phosphatase (LMW-PTP) with a small molecule inhibitor SPAA-2 | | Descriptor: | 2-[(1,3-benzothiazol-2-yl)amino]-2-oxoethane-1-sulfonic acid, Low molecular weight phosphotyrosine protein phosphatase | | Authors: | Wang, J, Zhang, Z.Y. | | Deposit date: | 2022-05-02 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Based Design of Active-Site-Directed, Highly Potent, Selective, and Orally Bioavailable Low-Molecular-Weight Protein Tyrosine Phosphatase Inhibitors.
J.Med.Chem., 65, 2022
|
|
7DJZ
 
 | | Crystal structure of SARS-CoV-2 Spike RBD in complex with MW01 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, MW01 heavy chain, ... | | Authors: | Wang, J, Jiao, S, Wang, R, Zhang, J, Zhang, M, Wang, M. | | Deposit date: | 2020-11-22 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Antibody-dependent enhancement (ADE) of SARS-CoV-2 pseudoviral infection requires Fc gamma RIIB and virus-antibody complex with bivalent interaction.
Commun Biol, 5, 2022
|
|
7DK0
 
 | | Crystal structure of SARS-CoV-2 Spike RBD in complex with MW05 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MW05 heavy chain, MW05 light chain, ... | | Authors: | Wang, J, Jiao, S, Wang, R, Zhang, J, Zhang, M, Wang, M. | | Deposit date: | 2020-11-22 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.199 Å) | | Cite: | Antibody-dependent enhancement (ADE) of SARS-CoV-2 pseudoviral infection requires Fc gamma RIIB and virus-antibody complex with bivalent interaction.
Commun Biol, 5, 2022
|
|
6C6A
 
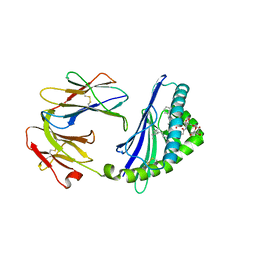 | | Structure of glycolipid aGSA[16,6P] in complex with mouse CD1d | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Zajonc, D.M, Wang, J. | | Deposit date: | 2018-01-18 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A molecular switch in mouse CD1d modulates natural killer T cell activation by alpha-galactosylsphingamides.
J.Biol.Chem., 294, 2019
|
|
4JFG
 
 | | Crystal structure of sfGFP-66-HqAla | | Descriptor: | CESIUM ION, Green fluorescent protein, quinolin-8-ol | | Authors: | Wang, J, Liu, X, Li, J, Zhang, W, Hu, M, Zhou, J. | | Deposit date: | 2013-02-28 | | Release date: | 2013-10-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Significant expansion of the fluorescent protein chromophore through the genetic incorporation of a metal-chelating unnatural amino acid.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
6C6E
 
 | | Structure of glycolipid aGSA[26,6P] in complex with mouse CD1d | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Zajonc, D.M, Wang, J. | | Deposit date: | 2018-01-18 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | A molecular switch in mouse CD1d modulates natural killer T cell activation by alpha-galactosylsphingamides.
J.Biol.Chem., 294, 2019
|
|
2A5J
 
 | | Crystal Structure of Human RAB2B | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related protein Rab-2B | | Authors: | Dong, A, Wang, J, Shen, Y, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Bochkarev, A, Park, H.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-06-30 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Crystal structure of human RAB2B
To be Published
|
|
2BAZ
 
 | |
4O0U
 
 | |
6J5S
 
 | | Crystal structure of human HINT1 mutant complexing with AP5A | | Descriptor: | BIS(ADENOSINE)-5'-PENTAPHOSPHATE, ETHANESULFONIC ACID, Histidine triad nucleotide-binding protein 1 | | Authors: | Wang, J, Fang, P, Guo, M. | | Deposit date: | 2019-01-11 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Second messenger Ap4A polymerizes target protein HINT1 to transduce signals in Fc epsilon RI-activated mast cells.
Nat Commun, 10, 2019
|
|
5GPH
 
 | |
6J53
 
 | | Crystal structure of human HINT1 complexing with ATP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Histidine triad nucleotide-binding protein 1 | | Authors: | Wang, J, Fang, P, Guo, M. | | Deposit date: | 2019-01-10 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Second messenger Ap4A polymerizes target protein HINT1 to transduce signals in Fc epsilon RI-activated mast cells.
Nat Commun, 10, 2019
|
|
6J58
 
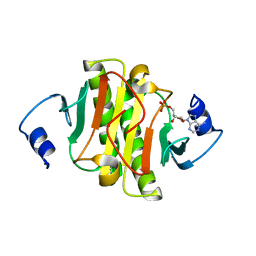 | | Crystal structure of human HINT1 complexing with AP4A | | Descriptor: | ADENOSINE MONOPHOSPHATE, Histidine triad nucleotide-binding protein 1 | | Authors: | Wang, J, Fang, P, Guo, M. | | Deposit date: | 2019-01-10 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.521 Å) | | Cite: | Second messenger Ap4A polymerizes target protein HINT1 to transduce signals in Fc epsilon RI-activated mast cells.
Nat Commun, 10, 2019
|
|
6J64
 
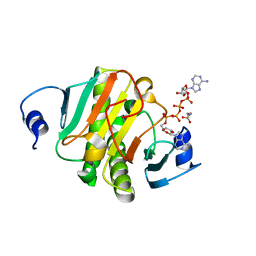 | | Crystal structure of human HINT1 mutant complexing with AP4A | | Descriptor: | 2-AMINOETHANESULFONIC ACID, BIS(ADENOSINE)-5'-TETRAPHOSPHATE, Histidine triad nucleotide-binding protein 1 | | Authors: | Wang, J, Fang, P, Guo, M. | | Deposit date: | 2019-01-14 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Second messenger Ap4A polymerizes target protein HINT1 to transduce signals in Fc epsilon RI-activated mast cells.
Nat Commun, 10, 2019
|
|
5HWT
 
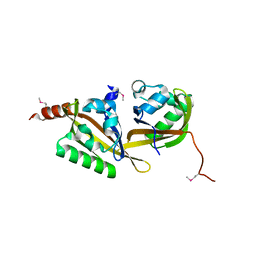 | | Crystal structure of apo-PAS1 | | Descriptor: | Sensor histidine kinase TodS | | Authors: | Hwang, J, Koh, S. | | Deposit date: | 2016-01-29 | | Release date: | 2016-03-02 | | Last modified: | 2017-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular Insights into Toluene Sensing in the TodS/TodT Signal Transduction System.
J. Biol. Chem., 291, 2016
|
|
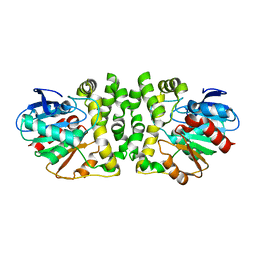
8HGU
 
 | |
5HWW
 
 | | Crystal structure of PAS1 complexed with 1,2,4-TMB | | Descriptor: | 1,2,4-trimethylbenzene, Sensor histidine kinase TodS | | Authors: | Hwang, J, Koh, S. | | Deposit date: | 2016-01-29 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Insights into Toluene Sensing in the TodS/TodT Signal Transduction System.
J. Biol. Chem., 291, 2016
|
|
