7Y3E
 
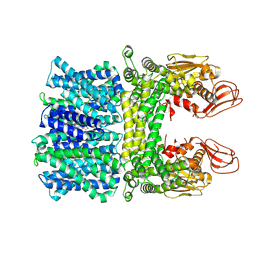 | |
8K5A
 
 | | The cryo-EM map of open TIEA-TIC complex | | Descriptor: | 15 kDa RNA polymerase-binding protein, DNA (29-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Zhang, K.N, Liu, Y, Chen, M, Wang, Y, Lin, W, Li, M, Zhang, X, Gao, Y, Gong, Q, Chen, H, Steve, M, Li, S, Zhang, K, Liu, B. | | Deposit date: | 2023-07-21 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | TIEA inhibits sigma70-dependent transcriptions, accelerates elongation speed and elevates transcription error
To Be Published
|
|
8K58
 
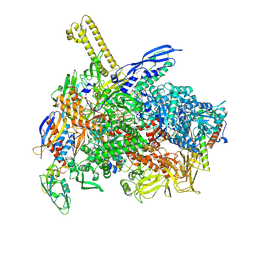 | | The cryo-EM map of close TIEA-TIC complex | | Descriptor: | 15 kDa RNA polymerase-binding protein, DNA (29-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Zhang, K.N, Liu, Y, Chen, M, Wang, Y, Lin, W, Li, M, Zhang, X, Gao, Y, Gong, Q, Chen, H, Steve, M, Li, S, Zhang, K, Liu, B. | | Deposit date: | 2023-07-21 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | TIEA inhibits Sigma70-dependent transcriptions, accelerates elongation speed and elevates transcription error
To Be Published
|
|
3JYN
 
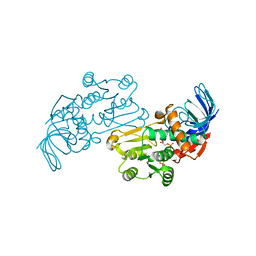 | | Crystal structures of Pseudomonas syringae pv. Tomato DC3000 quinone oxidoreductase complexed with NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Quinone oxidoreductase | | Authors: | Pan, X, Zhang, H, Gao, Y, Li, M, Chang, W. | | Deposit date: | 2009-09-22 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structures of Pseudomonas syringae pv. tomato DC3000 quinone oxidoreductase and its complex with NADPH
Biochem.Biophys.Res.Commun., 390, 2009
|
|
3JYL
 
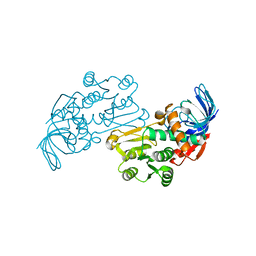 | | Crystal structures of Pseudomonas syringae pv. Tomato DC3000 quinone oxidoreductase | | Descriptor: | Quinone oxidoreductase | | Authors: | Pan, X, Zhang, H, Gao, Y, Li, M, Chang, W. | | Deposit date: | 2009-09-22 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of Pseudomonas syringae pv. tomato DC3000 quinone oxidoreductase and its complex with NADPH
Biochem.Biophys.Res.Commun., 390, 2009
|
|
3P6Y
 
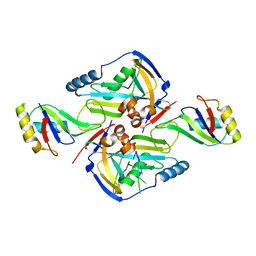 | | CF Im25-CF Im68-UGUAA complex | | Descriptor: | 5'-R(*UP*GP*UP*AP*A)-3', Cleavage and polyadenylation specificity factor subunit 5, Cleavage and polyadenylation specificity factor subunit 6 | | Authors: | Li, H, Tong, S, Li, X, Shi, H, Gao, Y, Ge, H, Niu, L, Teng, M. | | Deposit date: | 2010-10-11 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of pre-mRNA recognition by the human cleavage factor Im complex
To be Published
|
|
3P5T
 
 | | CFIm25-CFIm68 complex | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 5, Cleavage and polyadenylation specificity factor subunit 6 | | Authors: | Li, H, Tong, S, Li, X, Shi, H, Gao, Y, Ge, H, Niu, L, Teng, M. | | Deposit date: | 2010-10-11 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of pre-mRNA recognition by the human cleavage factor Im complex
To be Published
|
|
6IQI
 
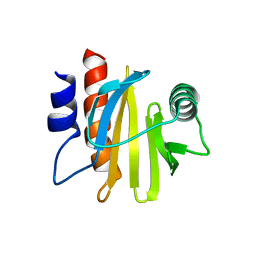 | | crystal structure of Arabidopsis thaliana Profilin 2 | | Descriptor: | Profilin-2 | | Authors: | Qiao, Z, Gao, Y. | | Deposit date: | 2018-11-08 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and computational examination of theArabidopsisprofilin-Poly-P complex reveals mechanistic details in profilin-regulated actin assembly.
J.Biol.Chem., 294, 2019
|
|
6M7V
 
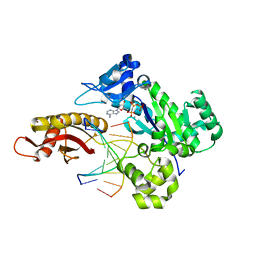 | | Human DNA polymerase eta extension complex with cdA at the -1 position | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(P*(02I)P*CP*TP*CP*AP*CP*AP*CP*T)-3'), DNA (5'-D(P*GP*TP*GP*TP*GP*AP*GP*T)-3'), ... | | Authors: | Weng, P, Gao, Y, Yang, W. | | Deposit date: | 2018-08-21 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.062 Å) | | Cite: | Bypassing a 8,5'-cyclo-2'-deoxyadenosine lesion by human DNA polymerase eta at atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6M7O
 
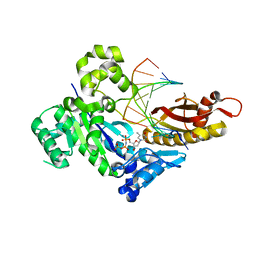 | | Human DNA polymerase eta ternary complex with Mn2+ and dTMPNPP oppositing cdA | | Descriptor: | 5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]thymidine, DNA (5'-D(*AP*TP*(02I)P*CP*TP*CP*AP*CP*AP*CP*T)-3'), DNA (5'-D(P*AP*GP*TP*GP*TP*GP*AP*G*(1FZ))-3'), ... | | Authors: | Weng, P, Gao, Y, Yang, W. | | Deposit date: | 2018-08-20 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Bypassing a 8,5'-cyclo-2'-deoxyadenosine lesion by human DNA polymerase eta at atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6IQJ
 
 | |
5BT1
 
 | | histone chaperone Hif1 playing with histone H2A-H2B dimer | | Descriptor: | HAT1-interacting factor 1, Histone H2A.1, Histone H2B.1 | | Authors: | Liu, H, Zhang, M, Gao, Y, Teng, M, Niu, L. | | Deposit date: | 2015-06-02 | | Release date: | 2016-10-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural Insights into the Association of Hif1 with Histones H2A-H2B Dimer and H3-H4 Tetramer
Structure, 24, 2016
|
|
6M7T
 
 | | Human DNA polymerase eta in a non-productive ternary complex with Ca2+ and dTTP oppositing cdA | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DNA (5'-D(*AP*GP*TP*GP*TP*GP*AP*G)-3'), ... | | Authors: | Weng, P, Gao, Y, Yang, W. | | Deposit date: | 2018-08-21 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Bypassing a 8,5'-cyclo-2'-deoxyadenosine lesion by human DNA polymerase eta at atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6M7U
 
 | | Human DNA polymerase eta in a non-productive ternary complex with Mg2+ and dTMPNPP oppositing cdA | | Descriptor: | 5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]thymidine, CHLORIDE ION, DNA (5'-D(*AP*GP*TP*GP*TP*GP*AP*G)-3'), ... | | Authors: | Weng, P, Gao, Y, Yang, W. | | Deposit date: | 2018-08-21 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Bypassing a 8,5'-cyclo-2'-deoxyadenosine lesion by human DNA polymerase eta at atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4GBV
 
 | | Crystal Structure of AMP complexes of Porcine Liver Fructose-1,6-bisphosphatase Mutant A54L with 1,2-ethanediol as Cryo-protectant | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase 1, ... | | Authors: | Honzatko, R.B, Gao, Y. | | Deposit date: | 2012-07-28 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Water Structure of the Central Hydrophobic Cavity of Mammalian Fructose-1,6-bisphosphatase: a Potential Thermodynamic Determinant of Allowed Quaternary States
To be published
|
|
4GBW
 
 | | Crystal Structure of AMP complexes of Porcine Liver Fructose-1,6-bisphosphatase Mutant A54L with 1,2-propanediol as Cryo-protectant | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase 1, ... | | Authors: | Honzatko, R.B, Gao, Y. | | Deposit date: | 2012-07-28 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Water Structure of the Central Hydrophobic Cavity of Mammalian Fructose-1,6-bisphosphatase: a Potential Thermodynamic Determinant of Allowed Quaternary States
To be published
|
|
4HGD
 
 | | Structural insights into yeast Nit2: C169S mutant of yeast Nit2 in complex with an endogenous peptide-like ligand | | Descriptor: | CACODYLATE ION, GLYCEROL, N-(4-carboxy-4-oxobutanoyl)-L-cysteinylglycine, ... | | Authors: | Liu, H, Qiu, X, Zhang, M, Gao, Y, Niu, L, Teng, M. | | Deposit date: | 2012-10-08 | | Release date: | 2013-07-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structures of enzyme-intermediate complexes of yeast Nit2: insights into its catalytic mechanism and different substrate specificity compared with mammalian Nit2
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HG3
 
 | | Structural insights into yeast Nit2: wild-type yeast Nit2 in complex with alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, CACODYLATE ION, GLYCEROL, ... | | Authors: | Liu, H, Qiu, X, Zhang, M, Gao, Y, Niu, L, Teng, M. | | Deposit date: | 2012-10-06 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structures of enzyme-intermediate complexes of yeast Nit2: insights into its catalytic mechanism and different substrate specificity compared with mammalian Nit2
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4H5U
 
 | | Structural insights into yeast Nit2: wild-type yeast Nit2 | | Descriptor: | CACODYLATE ION, GLYCEROL, Probable hydrolase NIT2 | | Authors: | Liu, H, Qiu, X, Zhang, M, Gao, Y, Niu, L, Teng, M. | | Deposit date: | 2012-09-18 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structures of enzyme-intermediate complexes of yeast Nit2: insights into its catalytic mechanism and different substrate specificity compared with mammalian Nit2
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HG5
 
 | | Structural insights into yeast Nit2: wild-type yeast Nit2 in complex with oxaloacetate | | Descriptor: | CACODYLATE ION, GLYCEROL, OXALOACETATE ION, ... | | Authors: | Liu, H, Qiu, X, Zhang, M, Gao, Y, Niu, L, Teng, M. | | Deposit date: | 2012-10-07 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structures of enzyme-intermediate complexes of yeast Nit2: insights into its catalytic mechanism and different substrate specificity compared with mammalian Nit2
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4NQ0
 
 | | Structural insights into yeast histone chaperone Hif1: a scaffold protein recruiting protein complexes to core histones | | Descriptor: | HAT1-interacting factor 1 | | Authors: | Liu, H, Zhang, M, He, W, Zhu, Z, Teng, M, Gao, Y, Niu, L. | | Deposit date: | 2013-11-23 | | Release date: | 2014-07-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into yeast histone chaperone Hif1: a scaffold protein recruiting protein complexes to core histones
Biochem.J., 462, 2014
|
|
4OQT
 
 | | LINGO-1/Li81 Fab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of Li81 Fab, ... | | Authors: | Pepinsky, R.B, Arndt, J.W, Quan, C, Gao, Y, Quintero-Monzon, O, Lee, X, Mi, S. | | Deposit date: | 2014-02-10 | | Release date: | 2014-05-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Structure of the LINGO-1-Anti-LINGO-1 Li81 Antibody Complex Provides Insights into the Biology of LINGO-1 and the Mechanism of Action of the Antibody Therapy.
J.Pharmacol.Exp.Ther., 350, 2014
|
|
7E3L
 
 | | Ultrapotent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 58G6 heavy chain, 58G6 light chain, ... | | Authors: | Guo, H, Li, T, Liu, F, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2021-02-09 | | Release date: | 2021-09-15 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Potent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants.
Nat Commun, 12, 2021
|
|
7E3K
 
 | | Ultrapotent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants | | Descriptor: | 13G9 heavy chain, 13G9 light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, H, Li, T, Liu, F, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2021-02-09 | | Release date: | 2021-09-15 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Potent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants.
Nat Commun, 12, 2021
|
|
3UNW
 
 | | Crystal Structure of Human GAC in Complex with Glutamate | | Descriptor: | GLUTAMIC ACID, Glutaminase kidney isoform, mitochondrial | | Authors: | DeLaBarre, B, Gross, S, Cheng, F, Gao, Y, Jha, A, Jiang, F, Song, J.J, Wie, W, Hurov, J. | | Deposit date: | 2011-11-16 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Full-length human glutaminase in complex with an allosteric inhibitor.
Biochemistry, 50, 2011
|
|
